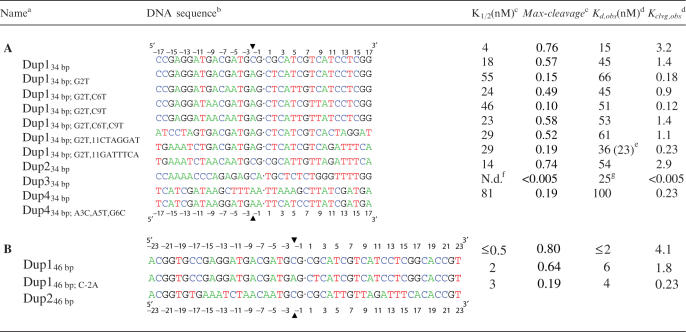
Table 1.
(A) 34 bp DNA duplexes; (B) 46 bp DNA duplexes and their thermodynamic parameters for binding and cleavage
aBase changes of only the 5′-half of the duplexes are indicated in the names of the duplexes. To keep the DNA duplexes palindromic, corresponding mutations were also introduced in the 3′-half.
bSequences of only one strand of the duplexes are shown from 5′ to 3′. All duplexes are palindromic. The center of the molecule is indicated (dot). The numbering of the bases is relative to the center of the duplex (see ruler) with the base immediately to the left having the position −1 and that to the right the position 1. The enzyme covalently attaches to the −2 position of each strand (arrowhead).
cBest fit parameters from Figure 1 and analogous titrations (see figure legend and Results section for details).
dKd,obs and Kclvg,obs are defined in the Results section. Except where indicated, values calculated from K1/2 and Kclvg,obs (see the Results section). Values for Kd,obs vary <2 two-fold, for Kclvg,obs < 50% in independent experiments (see the Materials and Methods section).
eValue in parenthesis measured in competition experiments (Figure 2).
fN.d.: not determined.