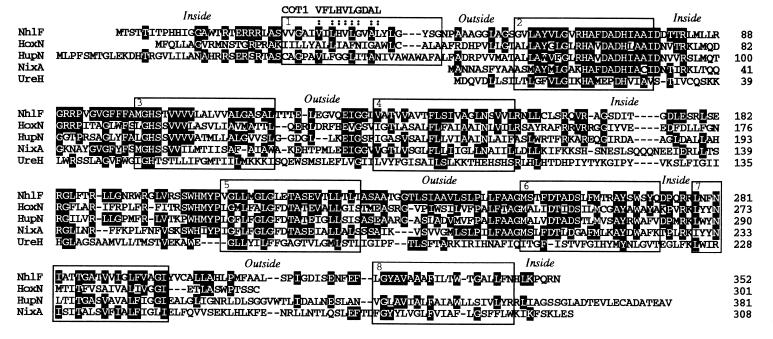
Figure 2.
Alignment of amino acid sequences among NhlF from R. rhodochrous J1 and homologous proteins. Amino acid sequences of NhlF from R. rhodochrous J1, HoxN from A. eutrophus (9), HupN from B. japonicum (18), NixA from H. pylori (19), and UreH from Bacillus sp. (20) were aligned by introducing gaps (hyphens) to achieve maximum homology. Residues in solid boxes indicate identical sequences. Putative transmembrane or membrane-associated domains (1–8) is enclosed by boxes. The orientation (inside, internal; outside, external) of the nonmembrane loop regions, predicted by the “positive inside rule” (21) is shown above the sequence. Highly conserved residues between NhlF and COT1 from S. cerevisiae (22) are marked with colons (:).