Figure 4.
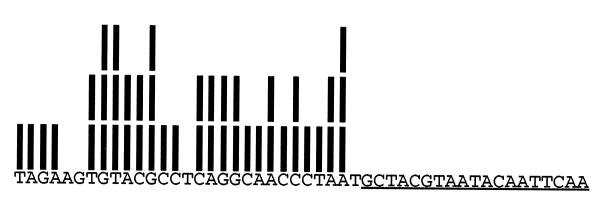
We developed a technique to examine the distribution of endpoints of genomic inserts that contain any short unique sequence (36). We isolated a set of library fragments that contain the Saccharomyces cerevisiae NDC1 gene sequence underlined above. Each bar above a nucleotide in the adjacent sequence means that nucleotide was adjacent to the T7 containing promoter in one of the 43 fragments examined. This limited analysis revealed 25 out of 29 possible endpoints! In our examination of five genes in four libraries, the largest “gap” that we found (i.e., stretch of nucleotides for which no endpoints were observed) is 9 nt. Assuming judicious choice of insert size, we are confident that every nucleic acid binding site for every protein is to be found in every library constructed by this method.