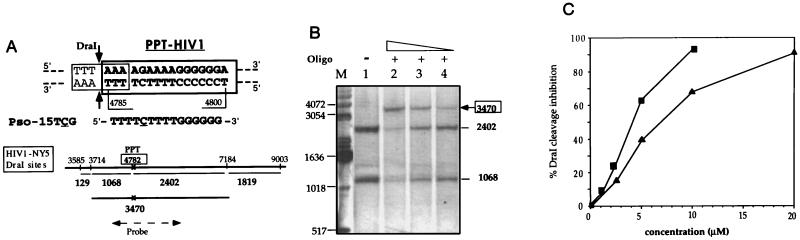
Figure 1.
DraI protection assay on naked genomic DNA. (A Upper) The 16-bp oligopurine·oligopyrimidine sequence we have chosen as a target for triple helix formation is indicated in boldface type. It is present in the HIV-1 genome and is called PPT. The PPT triplex site overlaps the DraI recognition sequence (thin line); the two arrows indicate the sites of DraI cleavage. The sequence of the psoralen-modified third strand is indicated. Underlined cytosine (C) indicates a 5 methyl cytosine. Pso- indicates a psoralen derivative attached to the 5′ phosphate (see Materials and Methods). (A Lower) DraI sites around the PPT triplex site (which is shown by a cross and boxed) present in the pol gene (HIV-NY5 sequence). sequence). The lengths of the fragments obtained after DraI cleavage are indicated. Location of the DNA probe (1902 bp long) used for Southern blot analysis is shown (broken arrow); the probe hybridizes with the 1068- and 2402-bp fragments over 897 and 1005 nt, respectively. (B) Naked genomic DNA (0.8 μg/μl) of U1 cells was irradiated (in the triplex-binding buffer) in the absence (lane 1) and in the presence of different concentrations of Pso-15TCG(po) (lane 2, 10 μM; lane 3, 5 μM; lane 4, 2.5 μM); the genomic DNA was then analyzed by DraI protection assay. Lane M, DNA marker (GIBCO/BRL). Fragments lengths (in base pairs) are indicated near the gel. (C) Quantitation of DraI cleavage inhibition at the PPT triplex site on the naked genomic DNA (as described in Fig. 2B) as a function of oligonucleotide concentration (▪, np; ▴, po). The ratio of the number of counts at the 3470-bp fragment to the sum of the 3470-, 2402-, and 1068-bp fragments are shown.