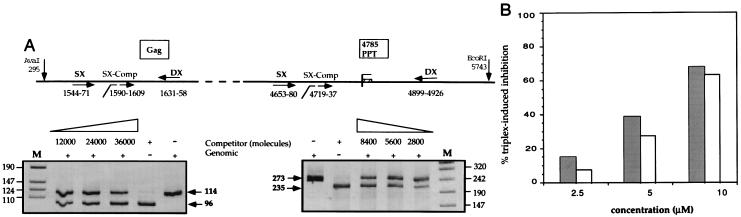
Figure 3.
Triplex-induced cross-link detection on genomic DNA by a competitive PCR-based assay. (A Upper) Schematic representation of the genomic HIV region, where the two primer pairs (Gag–SX, 5′-ATAATCCACCTATCCCAGTAGGAGAAAT-3′; and Gag–DX, 5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′; PPT–SX, 5′-CCCTACAATCCCCAAAGTCAAGGAGTAA-3′; and PPT-DX, 5′-GATCTCTGCTGTCCCTGTAATAAACCCG-3′) are located (converging arrows). For each primer pair, a competitor DNA fragment for quantitative PCR was constructed using DX primer and the internal SX–Comp primer [Gag–SX–Comp, 5′(Gag–SX)-CCTGGGATTAAATAAAATAG-3′; PPT–SX–Comp, 5′(PPT–SX)-CAGGTAAGAGATCAGGCTGA-3′, broken arrows] and quantitated. Before PCR amplification, genomic DNA was digested by EcoRI and AvaI which cut outside of the amplified regions as indicated. Nucleotide numbers refer to positions in HIV-1–NY5 (GenBank). (A Lower) Competitive PCR experiment for quantitation of the amount of cross-linked DNA molecules. Genomic DNA irradiated with 10 μM of Pso-15TCG(po) (see Fig. 1B, lane 2, which corresponds to the same sample analyzed by DraI protection assay) was quantitated by coamplification of a fixed amount of sample with increasing concentrations of competitor, as indicated near the gels. For each gel the genomic and competitor products are indicated. The results of quantitation obtained for these samples (20000 Gag molecules and 7400 PPT molecules) indicate that 63% of the DNA molecules are cross-linked. Lane M, DNA marker (VIII, Boehringer Mannheim). (B) Comparison of quantitations of the same sample after Pso-15TCG(po) treatment obtained by DraI protection assay (grey bars, corresponding to the samples of the experiment reported on Fig. 1) and by competitive PCR-based assay (white bars).