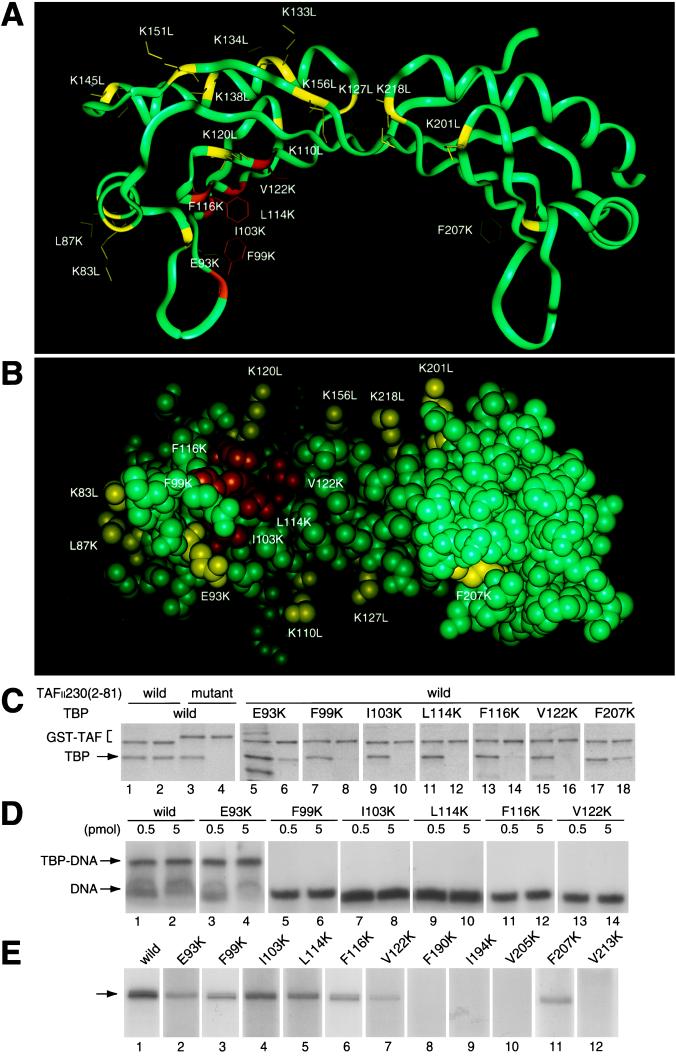
Figure 1.
dTAFII230-(2–81) binds to the concave surface of TBP. (A) Ribbon drawing of TBP viewed perpendicular toward the internal pseudodyad axis and (B) space-filling drawing viewed to the concave surface. The binding capability of TBP mutants to dTAFII230-(2–81) is indicated by colors. Mutations that cause a weak or no effect are shown by yellow (>50% activity of wild-type TBP), whereas mutations that cause serious defects are shown by orange (2–10%) or red (<2%). (C) TBP interaction with dTAFII230-(2–81). GST–dTAFII230-(2–81) was incubated with wild-type (lanes 1–4) or mutant (lanes 5–18) TBP. GST–dTAFII230-(2–81) with alanine substitutions in five contiguous residues (residues 24–28) (ref. 17) was used as a control (lanes 3 and 4). Input (odd lanes) and purified (even lanes) materials were analyzed by SDS/PAGE and visualized by Coomassie brilliant blue staining. (D) Analysis of TBP–DNA interactions by gel shift assay. Reaction mixtures contained 0.5 (odd lanes) or 5 (even lanes) pmol of wild-type (lanes 1 and 2) or mutant (lanes 3–14) TBP. (E) Analysis of basal transcription activity. Transcription activity was determined in a TBP-dependent reconstituted transcription system with 1 pmol of wild-type (lane 1) or mutant (lanes 2–12) TBP. The position of the accurately initiated transcript is indicated by an arrow.