Figure 5.
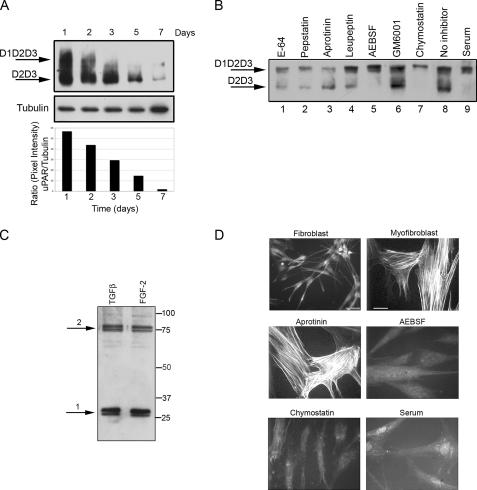
Proteases: AEBSF, chymostatin, and serum prevented uPAR cleavage and α-SMA stress fiber organization. (A) Time course of uPAR expression from 1 to 7 d. Cells were seeded in SSFM-TGFβ. At the specified time points, the cells were lysed and stored at −80°C. After the 7-d time point, cell lysates were detected for uPAR by Western blot using an uPAR antibody that detects total uPAR (D2). At 24 h, we detected the greatest amount of full-length uPAR, and by 7 d, uPAR expression was dramatically decreased. Tubulin expression functions as a control for protein loading. The ratio of the pixel densities (uPAR/tubulin) was calculated and graphed below. (B) Fibroblasts were grown for 24 in SSFM-TGFβ with E-64 (lane 1), pepstatin (lane 2), aprotinin (lane 3), leupeptin (lane 4), AEBSF (lane 5), GM6001 (lane 6), chymostatin (lane 7), no inhibitor (lane 8), or 10% FBS (lane 9). Cell lysates were immunodetected for total uPAR by Western blot. Cleaved uPAR (D2D3) was found in all cultures except those grown with AEBSF, chymostatin, or serum. (C) Based upon the impact of the inhibitors, cathepsin G is a candidate protease for cleaving uPAR. We detected cathepsin G in lysates of fibroblasts, grown for 24 h in either SSFM-TGFβ or SSFM-FGF. Arrow 1 denotes processed cathepsin G. Arrow 2 denotes cathepsin G complexed to α-1-anti-chymotrypsin. (D) Fibroblasts were grown for 72 h in SSFM-FGF (fibroblast), SSFM-TGFβ (myofibroblast), or SSFM-TGFβ plus aprotinin, AEBSF, chymostatin, or serum (10% FBS). Inhibitors were replaced every 24 h. α-SMA was detected by immunocytochemistry. Only the myofibroblast control and the aprotinin treated cell cultures incorporated α-SMA into stress fibers. Fibroblasts only (bar, 40 μm), myofibroblasts and inhibitor (bar, 20 μm). The images shown are representative of three experiments using two different tissue donors.