Abstract
Trehalose serves as a storage source of carbon and plays important roles under various stress conditions. For example, in many organisms trehalose has a critical function in preserving membrane structure and fluidity during dehydration/rehydration. In the yeast Saccharomyces cerevisiae, trehalose accumulates in the cell when the nutrient supply is limited but is rapidly degraded when the supply of nutrients is renewed. Hydrolysis of trehalose in yeast depends on neutral trehalase and acid trehalase (Ath1). Ath1 resides and functions in the vacuole; however, it appears to catalyze the hydrolysis of extracellular trehalose. Little is known about the transport route of Ath1 to the vacuole or how it encounters its substrate. Here, through the use of various trafficking mutants we showed that this hydrolase reaches its final destination through the multivesicular body (MVB) pathway. In contrast to the vast majority of proteins sorted into this pathway, Ath1 does not require ubiquitination for proper localization. Mutagenesis analyses aimed at identifying the unknown targeting signal revealed that the transmembrane domain of Ath1 contains the information sufficient for its selective sequestration into MVB internal vesicles.
INTRODUCTION
Trehalose, a nonreducing disaccharide in which two glucose molecules are connected in an α-1,1-glycosidic linkage, was first found in the ergot of rye in 1832 (Kopp et al., 1993). This sugar is widely distributed in various organisms, including bacteria, fungi, plants, insects, and invertebrates (Elbein, 1974). It was originally believed that trehalose served only as a sugar and energy reserve in the cell; however, it is now known that this molecule also functions as a structural component, plays a role in transport, and is involved in signaling (Elbein et al., 2003). But the most significant function of trehalose is in providing membrane protection against different kinds of stress conditions, such as heat, freezing, dehydration, anoxia, and nutrient limitation (Crowe et al., 1984).
Trehalose is quite common in yeast. In Saccharomyces cerevisiae, trehalose may constitute as much as 15–20% of its dry weight when growing in a stress environment. A strong correlation has been shown between trehalose content and stress resistance (Van Dijck et al., 1995). When yeast cells grow on rich carbon sources, they have a very low level of trehalose. In contrast, as they enter the stationary phase when nutrients are exhausted or during growth on nonfermentable carbon sources, the level of this disaccharide substantially increases. Conversely, when nutrients are resupplied, trehalose is rapidly mobilized (Nwaka and Holzer, 1998). The enzymes involved in synthesis and degradation of trehalose are the key regulators of these processes.
Currently the best studied pathway of trehalose biosynthesis is the one that catalyzes UDP-glucose and glucose-6-phosphate into trehalose by trehalose-6-phosphate synthase and trehalose-phosphate-phosphatase activities (Thevelein, 1984). On the other hand, the degradation of trehalose is catalyzed by trehalases. In S. cerevisiae, the hydrolysis of trehalose depends on two hydrolases: the cytoplasmic neutral trehalase (Nth1) and vacuolar localized acid trehalase (Ath1; Elbein et al., 2003). There have been many studies about the regulation and function of Nth1, but limited work has been done on Ath1. For example, there is still a lack of consensus concerning the vacuolar localization of Ath1. Wiemken and coworkers first found that Ath1 localizes in the vacuole after purification of this organelle by density gradient centrifugation (Keller et al., 1982). The low pH required for its maximal activity also suggests a localization within the vacuole (Mittenbuhler and Holzer, 1988). Recently, however, Jules et al. (2004) proposed that Ath1 is mainly distributed in the extracellular space. It is known that Nth1 is responsible for degrading intracellular trehalose and Ath1 for hydrolyzing extracellular trehalose (Nwaka et al., 1996). Yet it is not clear how Ath1 gets access to the extracellular substrate nor how this hydrolase transits to the vacuole, although the early secretory pathway has been shown to be involved (Harris and Cotter, 1988).
The late endosomes are the convergence point between endocytic (typically degradative) traffic from the cell surface and biosynthetic transport through the secretory pathway (Katzmann et al., 2002; Gruenberg and Stenmark, 2004; Babst, 2005; Slagsvold et al., 2006). Late endosomes undergo invagination of the limiting lipid bilayer to form internal vesicles and this process is used to sort lysosomal/vacuolar surface components from those destined to the interior of this organelle. Membrane proteins—both cargos destined for degradation and resident lysosomal/vacuolar enzymes destined to be liberated from the membrane—that are being transported to the lysosome/vacuole lumen are sequestered into these internal vesicles, whereas components targeted to the lysosome/vacuole surface are excluded from these structures and remain on the late endosome-limiting membrane. The resulting organelles are termed multivesicular bodies (MVBs) and fuse with lysosomes/vacuoles. During this event, the MVB-limiting membrane becomes part of the lysosome/vacuole surface whereas their content is released into the interior of this organelle, thereby delivering the different cargo molecules to their correct final location.
The formation of MVBs has been the object of intense study during the last few years, and part of the molecular machinery triggering this process has been in part unveiled (Babst, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006). The cytoplasmic domain(s) of most of the transmembrane components destined to the lysosome/vacuole lumen are mono-ubiquitinated in the late Golgi compartments or endosomal structures or at the plasma membrane (Katzmann et al., 2002; Reggiori and Pelham, 2002; Hettema et al., 2004). In the endosomes, these modified proteins are then recognized by the Vps27/Hrs-Hse1/STAM complex (sometimes referred to as endosomal sorting complex required for transport-0 [ESCRT-0]), which contains several ubiquitin-binding motifs, and this binding induces the recruitment of the ESCRT-I complex (Vps23/Tsg101, Vps28, and Vps37) to the endosomal membranes. This recruitment brings ESCRT-I in proximity to, and activates, the ESCRT-II complex (Vps22, Vps25, and Vps36), which receives ubiquitinated cargoes. ESCRT-II then promotes the assembly of the ESCRT-III complex (Vps2, Vps20, Vps24, and Snf7), leading to the concentration of the cargoes into a membrane domain that will invaginate inward. This chain of events also requires additional proteins including Bro1/Alix, Vta1, and Vps4, the latter being required for the recycling of the ESCRT complexes (Babst, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006; Russell et al., 2006).
To clarify the localization of Ath1 and to unravel its trafficking pathway, we used direct fluorescence microscopy and enzyme assays with purified vacuoles. Our results confirmed the location of Ath1 within the vacuole and further demonstrated that its vacuolar delivery requires the MVB pathway. Our analysis of Ath1 sorting into the MVB internal vesicles has led to the discovery that, unlike most other cargo proteins of the MVB pathway, this event is ubiquitin-independent and is mediated by the Ath1 transmembrane domain. These data provide insight into the molecular mechanism underlying the biosynthesis of Ath1, and additional information concerning the molecular mechanism underlying MVB biogenesis.
MATERIALS AND METHODS
Strains and Media
Strains used in this study are listed in Table 1. The YJH1 strain was generated by PCR-based integration of HA at the 3′ end of ATH1 in the BY4742 background using the method of Longtine et al. (1998). Strain YJH38 was made by replacing the entire coding region of the BSD2 gene with the Kluyveromyces lactis LEU2 gene that had been amplified with primers containing 40 base pairs of sequence identical to the flanking regions of BSD2 (Gueldener et al., 2002). Strain XGY5 was made by deleting the BSD2 gene with plasmid pDB25 (provided by Dr. Valerie Culotta, Johns Hopkins Bloomberg School of Public Health) in strain L3852.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| apm3Δ | BY4742 apm3Δ::KAN | ResGen |
| atg1Δ | BY4742 atg1Δ::KAN | ResGen |
| ath1Δ | BY4742 ath1Δ::KAN | ResGen |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | ResGen |
| end3Δ | BY4742 end3Δ::KAN | ResGen |
| fab1Δ | BY4742 fab1Δ::KAN | ResGen |
| FRY143 | SEY6210 vps4Δ::TRP1 pep4Δ::LEU2 | Cheong et al. (2005) |
| MHY623 | MATα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 doa4-Δ1::LEU2 | Papa and Hochstrasser (1993) |
| tul1Δ | BY4742 tul1Δ::KAN | ResGen |
| vps2Δ | BY4742 vps2Δ::KAN | ResGen |
| vps4Δ | BY4742 vps4Δ::KAN | ResGen |
| vps22Δ | BY4742 vps22Δ::KAN | ResGen |
| vps23Δ | BY4742 vps23Δ::KAN | ResGen |
| vps27Δ | BY4742 vps27Δ::KAN | ResGen |
| XGY5 | MATα his3-Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 bsd2Δ::LEU2 | This study |
| YJH1 | BY4742 ATH1-HA::HIS3 | This study |
| YJH35 | FRY143 ATH1-HA::KAN | This study |
| YJH38 | BY4742 tul1Δ::KAN bsd2Δ::LEU2 | This study |
S. cerevisiae cultures were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% glucose), SMT-URA (0.67% yeast nitrogen base, 2% trehalose, amino acids without uracil, and vitamins), or SMD-URA (0.67% yeast nitrogen base, 2% glucose, amino acids without uracil, and vitamins) media. Cells expressing endogenous Ath1-HA or green fluorescent protein (GFP)-Ath1 were grown to stationary phase to induce expression of ATH1.
Plasmids
The open reading frame together with the terminator of ATH1 was amplified by the PCR from S. cerevisiae genomic DNA of strain BY4742 and then digested with MfeI/BamHI. Plasmid pPEP12416 (described in Reggiori et al., 2000) was digested with EcoRI/BamHI to excise the PEP12 gene, and the resulting vector was ligated with the above digested ATH1 PCR product to create the pGFPATH1 plasmid, expressing GFP-Ath1 under the control of a constitutively active TPI1 promoter. To generate N-terminally truncated Ath1, the PCR product of the ATH1 gene lacking the first 45-amino acid coding sequence was digested with MfeI/BamHI and ligated into pPEP12416 between the EcoRI/BamHI sites as described above to create the pGFPATH1ΔN plasmid. To generate a C-terminally truncated Ath1, a fragment encoding GFP fused with the first 69 amino acids of Ath1 plus a stop codon was PCR-amplified from the previously generated plasmid pGFPATH1 and digested with HindIII/BamHI and then cloned into the same sites in pGFPATH1 to generate pGFPATH1ΔC. To make a GFP-fused transmembrane domain of Ath1, a fragment including sequences encoding the GFP-fused Ath1 transmembrane domain region plus a stop codon was PCR-amplified from template pGFPATH1ΔN and digested with and then ligated into the HindIII/BamHI sites on pGFPATH1ΔN to generate pGFPATH1TM. To make the pPromATH1GFPATH1 construct with the endogenous ATH1 promoter, a 500-base pair segment from the promoter region of ATH1 was PCR-amplified from genomic DNA and digested with XhoI/HindIII and exchanged with the TPI1 promoter on the plasmid pGFPATH1.
To make single K27R or K37R, or double K27,37R mutations in Ath1, we took advantage of an AgeI site located between lysines 27 and 37. A partial N-terminal ATH1 fragment was PCR amplified from the pGFPATH1 plasmid using primers that introduce an A-to-G point mutation at nucleotide 80, which changes lysine at position 27 into arginine. The PCR product was digested with Bsu36I/AgeI and ligated into plasmid pGFPATH1 digested with the same enzymes, generating pGFPATH1K27R. Additional primers were used to amplify a fragment of ATH1 with a K37R mutation, which was digested with AgeI/BamHI and ligated into the same sites in pGFPATH1 or pGFPATH1K27R to create the pGFPATH1K37R and pGFPATH1K27,37R plasmids. To make pGFPATH1K2R and pGFPATH1K2,27,37R plasmids, we used the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to generate the K2R mutation in the pGFPATH1 and pGFPATH1K27,37R plasmids. Polar amino acid mutations in the transmembrane domain of Ath1 were made by the SOEing PCR method (Horton et al., 1990). Primers included altered sequences to amplify fragments of the ATH1 gene with mutations of N49V, S50A, T65V, and Y68F using template plasmids, pGFPATH1 and pGFPATH1ΔN. The PCR products of the mutated ATH1 and ATH1ΔN were inserted into the plasmids described above to replace the wild-type ATH1 and ATH1ΔN segments. The corresponding gene products are referred to as GFP-Ath1polarmut and GFP-Ath1ΔNpolarmut. DNA sequencing was used to verify all of the introduced point mutations.
The plasmid YEp112 (pHA-Ub; Hochstrasser et al., 1991) was generously provided by Dr. Mark Hochstrasser (Yale University).
Fluorescence Microscopy
Cells expressing GFP-fused chimeras were grown in SMD-URA medium to midlogarithmic (log) phase or stationary phase. The vacuolar membrane was labeled with FM 4-64 (Molecular Probes, Eugene, OR) by incubating cells in medium containing 20 μM FM 4-64 at 30°C for 15 min and then washing with YPD medium once and incubating for another 30 min. Fluorescence signals were visualized with a DeltaVision Spectris fluorescence microscope (Applied Precision, Issaquah, WA). The images were captured with a CoolSnap camera and deconvolved using SoftWoRx software (Applied Precision).
Vacuole Preparation and Enzyme Assays
Wild-type and ath1Δ cells were grown to stationary phase in YPD and vacuoles from each strain were isolated as described previously (Haas, 1995; Hutchins and Klionsky, 2001). The acid trehalase, α-mannosidase, α-glucosidase, and NADPH cytochrome c reductase assays were performed as described (Opheim, 1978; Johnson et al., 1987; Alizadeh and Klionsky, 1996). All enzyme assays were performed on lysates loaded onto the ficoll gradient and on the isolated vacuole fraction. Vacuolar Ath1 activity was normalized relative to the recovery of α-mannosidase in the vacuole fraction.
Subcellular Fractionation and Immunoblot
Cells from strain YJH1 (BY4742, Ath1-HA) were grown in YPD medium to stationary phase and incubated in 0.1 M Tris-HCl, pH 9.4, containing 30 mM 2-mercaptoethanol at room temperature for 20 min. Cells were collected by centrifugation, and the cell pellet was resuspended in spheroplasting medium (1.2 M sorbitol, 20 mM Tris-HCl, pH 7.5, 40 μg/ml yeast lytic enzyme) and incubated at 30°C for 30 min with gentle shaking. The spheroplasts were then subjected to differential lysis in PS200 lysis buffer (20 mM PIPES, pH 6.8, 200 mM sorbitol, 5 mM MgCl2) containing Complete EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN) and 1 mM phenylmethylsulfonyl fluoride (PMSF). After a preclearing step at 300 × g in an Eppendorf 5415D microcentrifuge for 5 min at 4°C, the lysate was subjected to low-speed centrifugation at 13,000 × g for 5 min at 4°C. The low-speed supernatant (S13) and pellet (P13) fractions were separated for further analysis.
For biochemical characterization of Ath1 membrane association, the P13 fraction was resuspended in equal volumes of PS0 buffer (0.2 M PIPES-NaOH, pH 7.8) containing 1% Triton X-100 (TX-100), 0.1 M Na2CO3, pH 11, or 1.0 M KCl. After a 5-min incubation at room temperature, the treated lysates were centrifuged at 13,000 × g for 5 min at 4°C to separate supernatant and pellet fractions.
For immunoblotting, antisera against GFP, Pho8, and HA were purchased from Covance Research Products, (Berkeley, CA), Molecular Probes and Santa Cruz Biotechnology (Santa Cruz, CA), respectively.
Protease Protection Assay
After subcellular fractionation, the low-speed pellet fraction (P13) was treated with 50 μg/ml proteinase K (in lysis buffer PS200) alone, 1% TX-100, or both. After incubation on ice for 15 min, lysates were subjected to 10% trichloroacetic acid (TCA) precipitation and processed for Western blot.
Immunoprecipitation
Yeast cells were grown to log phase, and 30 OD600 units (1 U is equivalent to 1 ml of cells at OD600 = 1.0) of cells were collected and lysed by glass beads in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% TX-100, 1 mM PMSF, and protease inhibitor cocktail) with addition of 5 mM N-ethylmaleimide (NEM). The lysate was incubated with protein A Sepharose and anti-HA or anti-GFP antibody (Santa Cruz Biotechnology) at 4°C for 4 h. The Sepharose was washed three times in lysis buffer with 5 mM NEM and finally eluted in SDS loading buffer. Samples were subjected to SDS-PAGE and immunoblotted with anti-GFP, anti-HA, anti-Cps1 (generously provided by Dr. Scott Emr, Cornell University; Cowles et al., 1997), or anti-ubiquitin (Zymed Laboratories, South San Francisco, CA/Invitrogen, Carlsbad, CA) antibodies or antiserum.
Endoglycosidase H Treatment
Wild-type cells harboring the pGFPAth1 plasmid were grown to midlog phase and 5 OD600 units of cells were collected and subjected to TCA precipitation. The pellet fraction was dried and resuspended in 100 μl elution buffer (0.1 M Tris-HCl, pH 7.5, 1% SDS, 1% 2-mercaptoethanol) by sonication. The sample was heated at 95°C for 5 min and 900 μl of Endoglycosidase H (endo H) buffer (0.15 M citric acid, pH 5.5) were added. After a quick spin, the supernatant fraction was split into two tubes. PMSF (2 mM final concentration) and 1× protease inhibitors cocktail were added to each tube and mixed by vortex. Endo H (10 mU) was added to one of the two tubes, and both were incubated at 37°C overnight. The mixture was then TCA-precipitated, and the pellet fraction was resuspended in 50 μl sample buffer and subjected to immunoblotting.
RESULTS
Ath1 Is Localized in the Interior of the Vacuole in S. cerevisiae
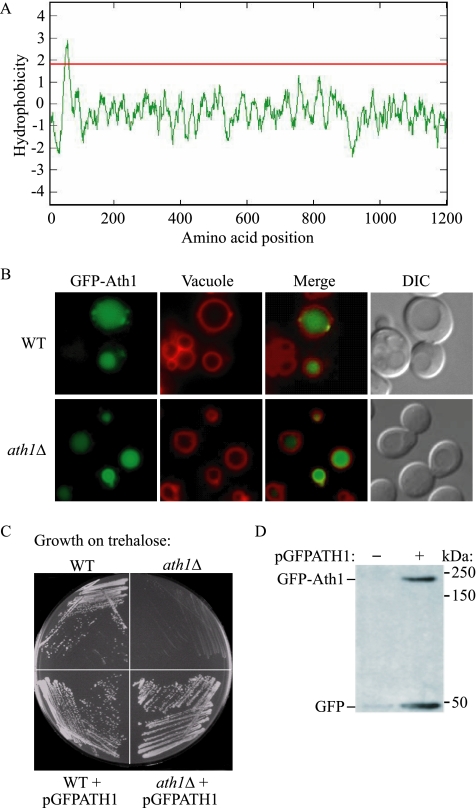
To investigate the location of Ath1 in S. cerevisiae and to identify its biosynthetic pathway, we fused the GFP to Ath1 and directly monitored its localization by live cell fluorescence microscopy. According to its amino acid sequence, Ath1 is predicted to have a transmembrane domain near the amino terminus based on Kyte-Doolittle hydropathy analysis (Kyte and Doolittle, 1982); the majority of the protein is present in the carboxyl-terminal tail (Figure 1A). Accordingly, it is highly probable that the catalytically active domain of Ath1 is within its C terminus. Thus, we fused GFP to the N terminus of Ath1 in order to minimize potential affects on the function of the protein. Importantly, this tagging strategy has been used with other yeast vacuolar hydrolases with a similar topology, such as Cps1 and Phm5, and has been shown to not affect their trafficking and localization (Odorizzi et al., 1998; Reggiori and Pelham, 2001). We generated a construct, pGFPATH1, which expresses an N-terminal GFP-fused Ath1 under a strong TPI1 promoter and transformed it into wild-type and ath1Δ strains. Cells were incubated with FM 4-64, a fluorescent red lipid dye that specifically stains the yeast vacuole membrane (Vida and Emr, 1995), and imaged by fluorescence microscopy. Most of the GFP signal was seen enclosed within the red ring-shaped vacuole membrane, which indicated localization of Ath1 in the vacuolar lumen (Figure 1B).
Figure 1.
Ath1 is localized in the vacuolar lumen in S. cerevisiae. (A) A hydropathy plot was created by the Kyte-Doolittle algorithm (Kyte and Doolittle, 1982) based on the amino acid sequence of Ath1, and one transmembrane domain was predicted at amino acids 51–69. Peaks with scores greater than 1.8 (line) indicate a possible transmembrane region. (B) Wild-type (WT; BY4742) and ath1Δ strains were transformed with a plasmid (pGFPATH1) encoding a GFP-Ath1 fusion protein controlled by a constitutively active, strong TPI1 promoter. Yeast cells were grown in SMD-URA medium to log phase, treated with FM 4-64 to stain the vacuole and observed with a fluorescence microscope as described in Materials and Methods. DIC, differential interference contrast. (C) WT, ath1Δ, or the same strains harboring the pGFPATH1 plasmid were streaked on SMT-URA medium in which trehalose is the sole carbon source. The plate was grown at 30°C for 7 d. (D) The ath1Δ strain harboring either the pRS416 empty vector (−) or pGFPATH1 (+) plasmid were collected at log phase and whole cell extracts were subjected to SDS-PAGE. GFP or GFP-Ath1 were detected by immunoblot probed with monoclonal anti-GFP antibody. Molecular mass (kDa) of protein standards is indicated on the right side of the blot.
Functionality of the fusion chimera GFP-Ath1 was confirmed by testing its ability to complement the growth defect of ath1Δ cells in medium with trehalose as the sole carbon source, and expression was confirmed by Western blot probed with GFP antibody (Figure 1, C and D; Nwaka et al., 1996). On the Western blot, two protein bands were detected in cells harboring the pGFPATH1 plasmid but not those with an empty vector. One of the two bands migrated at ∼200 kDa, corresponding to the predicted size of the fusion protein, GFP-Ath1, whereas the other band with a 25-kDa molecular mass represented free GFP. The generation of free GFP was dependent on vacuolar proteinase A, the product of the PEP4 gene, because no free GFP could be seen in a pep4Δ background (see Figure 4C and our unpublished results). This finding suggested that the cleavage of GFP from the fusion protein occurred within the vacuole lumen. To eliminate the possibility of artifacts resulting from overexpression of the protein, we also checked the intracellular localization and functionality of GFP-Ath1 expressed under its endogenous promoter. The results were essentially the same as with the overexpressed protein; however, the fluorescence signal was substantially reduced (see Figure 3B and our unpublished results).
Figure 4.
Ath1 is a glycosylated, type II transmembrane protein. (A) A whole cell extract from wild-type cells harboring the pGFPAth1 plasmid was prepared and treated without (−) or with (+) endoglycosidase H as described in Materials and Methods. Samples were resolved by SDS-PAGE and subjected to Western blot probed with anti-GFP, anti-Prc1, and anti-Ape1 antibody or antisera (faster migrating species corresponding to free GFP were not detected). Protein bands of precursor Ape1 (prApe1) and mature Ape1 (mApe1) are indicated. (B) Schematic diagram of the localization and cleavage patterns of the GFP-Ath1 chimera in wild-type (WT), vps4Δ, and pep4Δ vps4Δ strains. The predicted molecular mass of the GFP-containing species is indicated. (C) Cells from wild-type (WT), vps4Δ, and pep4Δ vps4Δ strains were transformed with the pRS416 empty vector (−) or the pGFPAth1 plasmid (+). Whole cell extracts were analyzed by immunoblotting with anti-GFP antibody. The asterisks indicate nonspecific bands.
Figure 3.
Ath1 is delivered to the vacuole through the MVB pathway. GFP-Ath1 was either expressed under the TPI1 promoter (A) or under its endogenous promoter (B) in wild-type (WT), atg1Δ, apm3Δ, vps4Δ, vps27Δ, and end3Δ strains. Cells overexpressing GFP-Ath1 were grown in SMD-URA medium to log phase, whereas cells expressing the endogenous level of GFP-Ath1 were grown to stationary phase to induce Ath1 expression. Vacuoles were labeled with FM 4-64, and GFP-Ath1 localization was observed with a fluorescence microscope.
Recently Jules et al. (2004) showed that more than 90% of Ath1 activity is detected in the extracellular fraction. This result was dependent on the use of a modified trehalase assay using intact cells treated with sodium fluoride to prevent uptake of glucose generated from hydrolysis of trehalose; the trehalase assay measures free glucose and cellular uptake would result in the appearance of a lower level of extracellular product. Their findings conflict with previously published data (Keller et al., 1982) and our fluorescence data presented here. To address this discrepancy we isolated purified vacuoles (Haas, 1995) to directly monitor whether the activity of Ath1 was present within the vacuole fraction.
Approximately 53% of the total Ath1 activity from wild-type cells (set to 100%) was recovered in the vacuolar fraction (Figure 2A). As a negative control, ath1Δ cells, which should have no acid trehalase activity, yielded 23 and 12% trehalase activity in the total and vacuole fractions, respectively (Figure 2A). The efficiency of recovery of vacuoles and the purity of the vacuole fraction were examined by measuring α-mannosidase (vacuole marker), α-glucosidase (cytosol marker), and NADPH cytochrome c reductase (endoplasmic reticulum [ER] marker) activities (Figure 2B). Approximately 42% of the total vacuoles were recovered, and the contamination from cytosol and microsomes was ∼2.5 and 6%, respectively. Together with the fluorescence microscopy, these data indicate that Ath1 is primarily localized to the vacuole.
Figure 2.
Acid trehalase activity is primarily localized within the vacuole. (A) Vacuoles from the same amount of cells of the wild-type (WT; BY4742) and ath1Δ strains were prepared and assayed as described in Materials and Methods. Acid trehalase activity from the total cell lysate (Total) as well as the vacuole fraction (Vacuole) was measured. Final vacuolar Ath1 activity was calculated as the activity present in the vacuole fraction divided by the level of vacuole recovery according to the percentage of α-mannosidase in the vacuole fraction shown in B. The wild-type total Ath1 activity was set at 100%, and the other values were normalized accordingly. (B) Enzyme assays of α-mannosidase, α-glucosidase, and NADPH cytochrome c reductase were carried out as described in Materials and Methods. The percentages of enzyme activity in the vacuole fraction compared with the total loaded cell lysate from both wild-type and ath1Δ strains were plotted. All experiments were repeated three times independently. Error bar, SD among the repeats.
Ath1 Is Targeted to the Vacuole via the MVB Pathway
It is known that trehalose is distributed on both sides of the plasma membrane and can be found in the cytosol (Keller et al., 1982; Elbein et al., 2003). In contrast, it is not clear where Ath1 hydrolyzes its substrate. There are currently two models that propose the site of Ath1 activity: The first model is that Ath1 is transported to the plasma membrane where it binds trehalose located on the extracellular surface of the membrane. Both trehalose and trehalase are subsequently internalized via endocytosis and transported to the vacuole where the enzyme is activated and trehalose is hydrolyzed into glucose. In the second model, Ath1 is directly targeted to the vacuole similar to other vacuolar resident enzymes; trehalose is delivered to the vacuole through endocytosis, transport via a vacuole membrane transporter, and/or autophagy and is degraded within the vacuole lumen (Nwaka and Holzer, 1998). To determine the site of action of Ath1, we decided to examine the trafficking route of this enzyme.
In yeast, proteins can be sorted to the vacuole through several different transport pathways including the following: 1) The alkaline phosphatase (Alp) pathway delivers proteins directly from the trans-Golgi network (TGN) to the vacuole; the carboxypeptidase Y (CpY) pathway routes proteins from the TGN to endosomes and then to the vacuole; the MVB pathway involves movement of membrane proteins from the TGN to intralumenal endosomal vesicles (termed multivesicular bodies); the cytoplasm to vacuole targeting (Cvt)/autophagy pathway delivers proteins directly from the cytoplasm to the vacuole (Felder et al., 1990; Klionsky et al., 1992; Marcusson et al., 1994; Cowles et al., 1997; Odorizzi et al., 1998). The APM3 (Alp pathway), VPS4 (CpY and MVB pathways), and ATG1 (Cvt and autophagy pathways) genes encode components necessary for these pathways (Matsuura et al., 1997; Zahn et al., 2001; Avaro et al., 2002). If these genes are deleted, the corresponding pathways will be blocked. Vps4 is required for both the CpY and MVB pathways. This protein is a member of the class E Vps proteins, which are involved in invagination and formation of the intralumenal MVB vesicles, and vps4Δ mutant cells have one enlarged abnormal late endosome structure, called the class E compartment, in which cargo proteins are accumulated (Raymond et al., 1992; Odorizzi et al., 1998). However, the vps4Δ mutant has different phenotypes for cargoes of the CpY and MVB pathways. Vacuolar hydrolases using the CpY pathway are generally soluble proteins, which are not internalized into MVB vesicles. These proteins are partially secreted and mostly accumulate in the class E compartment in vps4Δ mutant cells (Babst et al., 1997). In contrast, cargoes of the MVB pathway are those integral membrane proteins that are normally selectively internalized into MVB vesicles and therefore accumulate on the limiting membrane of the abnormal late endosome as well as on the vacuole membrane when Vps4 function is compromised (Reggiori and Pelham, 2001).
To examine the vacuolar sorting pathway of Ath1, we transformed the pGFPATH1 plasmid into apm3Δ, vps4Δ, and atg1Δ strains and examined GFP-Ath1 localization. As shown in Figure 3A, in apm3Δ and atg1Δ cells, GFP-Ath1 was localized inside the vacuole lumen, the same as in wild-type cells, whereas in vps4Δ cells the green fluorescence signal was totally mislocalized on the surface of the class E compartment and the vacuole-limiting membrane. Vps4 is required for disassembly of the ESCRT complexes that are involved in the MVB pathway. We decided to extend our analysis by examining additional mutants representative of each of the ESCRT complexes to verify that the defect in localization observed in the vps4Δ strain was not specific to this mutant. Mutants defective in the function of Vps27 (ESCRT-0), Vps23 (ESCRT-I), Vps22 (ESCRT-II), and Vps2 (ESCRT-III) displayed mislocalization of GFP-Ath1 similar to that observed in the vps4Δ strain (Figure 3A and our unpublished results). These changes in localization suggested that the vacuolar targeting of Ath1 depends on the MVB pathway.
Although our data suggest that Ath1 enters the MVB pathway, there are still two possible routes through which this could occur. Ath1 could be either directly sorted into the MVB pathway at the TGN or first secreted to the plasma membrane by exocytosis and then internalized by endocytosis. To distinguish between these two possibilities, GFP-Ath1 was expressed in an endocytosis-deficient mutant, end3Δ, in which the internalization step of endocytosis is defective (Raths et al., 1993); Ath1 should be blocked at the plasma membrane in end3Δ cells if it goes through the exocytic and endocytic pathways en route to the vacuole. Alternatively, if Ath1 is directly sorted into the MVB pathway, there would not be any effect on Ath1 localization in end3Δ cells. We found that GFP-Ath1 was still localized in the lumen of the vacuole in the end3Δ mutant (Figure 3A). Similar results were obtained for GFP-Ath1 expressed under the control of its endogenous promoter (Figure 3B). This result eliminated the possibility that Ath1 is delivered to the plasma membrane before reaching the vacuole and thus argues against the model that Ath1 binds its substrate, trehalose, on the plasma membrane.
Ath1 Is a Glycosylated, Type II Transmembrane Protein
Detection of GFP-Ath1 with GFP antibody revealed a sharp band migrating at ∼200 kDa (Figure 4A). After treatment with endo H, which can remove N-linked glycans, a smaller band with a molecular mass around 160 kDa appeared and the higher band disappeared (Figure 4A). As controls for the endo H treatment we examined CpY (Prc1), which transits through the secretory pathway and undergoes glycosyl modification, and aminopeptidase I (Ape1), which is delivered to the vacuole independent of secretory pathway transit and is not glycosylated (Klionsky et al., 1992). Prc1 showed the expected change in molecular mass after enzyme treatment, whereas Ape1 was unaffected. These results indicated that Ath1 is glycosylated in a pattern that is typical of vacuolar proteins; it is not heterogeneously and extensively glycosylated as seen with proteins such as invertase that are secreted (Hong et al., 1996). Normally, protein glycosylation occurs in the ER and Golgi lumen, so the glycosylation of Ath1 also suggests that it transits through the early secretory pathway. Conversely, the absence of extensive glycosylation suggested that the protein is unlikely to be secreted. These data further supported our finding that Ath1 is transported through the MVB pathway, but bypasses the plasma membrane.
The MVB sorting of Ath1 implied that it is a membrane protein, because the invaginated vesicles in the late endosome only internalize membrane proteins. Moreover, according to the amino acid sequence of Ath1, a transmembrane domain is predicted near the N terminus, encompassing amino acids 51–69 (Figure 1A). This feature, together with its vacuolar trafficking pattern, is similar to that seen with Cps1, Phm5, and Atg15, vacuolar membrane proteins that utilize the MVB pathway (Katzmann et al., 2001; Reggiori and Pelham, 2001; Epple et al., 2003), which implies that Ath1 is likely also a type II transmembrane protein. To determine the topology of Ath1, we checked the cleavage pattern of Ath1 in wild-type, vps4Δ, and vps4Δ pep4Δ cells. Pep4 is directly or indirectly responsible for the processing of most vacuolar protein precursors. Ath1 activity is dependent on Pep4 (Harris and Cotter, 1987), but it is unclear whether Ath1 is proteolytically processed or where its cleavage site is located. If Ath1 is a type II transmembrane protein, its N terminus would be cytosolic and its C terminus would be located within the ER lumen before its final delivery into the vacuole. As shown in Figure 4B, in wild-type cells, after vacuolar delivery via the MVB pathway GFP-Ath1 will be present within the vacuole lumen where it will be fully accessible to proteases; the GFP moiety may be cleaved, so that a band corresponding to free GFP can be detected. In vps4Δ cells, however, Ath1 was mislocalized to the vacuole-limiting membrane, so that only the portion facing the vacuole lumen would be susceptible to cleavage; in this case free GFP should not be generated. On the other hand, if any site in the lumenal part of the protein was processed by vacuolar proteases in the vps4Δ mutant, we should be able to detect a GFP-fused protein band, which is larger than free GFP but smaller than full-length GFP-Ath1. In vps4Δ pep4Δ cells, even the lumenal part of the protein cannot be cleaved because of lack of Pep4 activity, so neither free GFP nor any intermediate-sized GFP fusion protein should be detected. Finally, if Ath1 has the opposite topology to that predicted, that is with the N terminus within the vacuole lumen, free GFP should be detected in both wild-type and vps4Δ cells.
Our results were consistent with the cleavage patterns expected for a type II integral membrane protein (Figure 4C). Whole cell extracts of various strains were resolved by Western blot and probed with GFP antibody. In the wild-type strain we saw an ∼25-kDa band corresponding to free GFP, whereas we detected only an ∼37-kDa band in vps4Δ cell extracts, which is predicted to correspond to free GFP plus the cytosolic and transmembrane domains of the Ath1 protein; free GFP or an intermediate-sized band was not seen in vps4Δ pep4Δ cells (Figure 4C), and only full-length GFP-Ath1 was detected in this background. In wild-type and vps4Δ cells, full-length GFP-Ath1 was also detected, but at a reduced level compared with the vps4Δ pep4Δ cells. These results further suggested that the GFP-Ath1 fusion protein is indeed targeted to the vacuole because the cleavage of GFP was dependent on the activity of the vacuolar hydrolase, Pep4. These results suggested that Ath1 is a type II transmembrane protein with the N terminus facing the cytosol.
To confirm the above results, we applied a biochemical analysis to examine the Ath1 membrane association and topology. Ath1 was chromosomally tagged with 3xHA at the C termimus; the Ath1-HA fusion protein was found to be functional, because it allowed growth of cells on trehalose medium (our unpublished results). Expression of Ath1-HA was detected by immunoblot using antiserum against HA, and only one protein band representing the fusion protein was seen in this strain, but not in wild-type cells in which Ath1 was not tagged with HA (Figure 5A). The presence of intact Ath1-HA suggested that no cleavage occurred at the C terminus, which would have removed the HA epitope. Subcellular fractionation experiments were performed to determine the membrane association of Ath1-HA. Spheroplasts were prepared and osmotically lysed as described in Materials and Methods. The cell lysate was separated into low-speed supernatant (S13) and pellet (P13) fractions by centrifugation at 13,000 × g. As shown in Figure 5A, Ath1-HA was recovered in the P13 fraction, which is known to contain the vacuole (Rieder and Emr, 2000). Cytosolic Pgk1 was recovered exclusively in the supernatant fraction, indicating efficient lysis of the spheroplasts and separation of the soluble and pelletable fractions. To determine whether Ath1 is a transmembrane protein, we chromosomally tagged the Ath1 C terminus with a 3xHA epitope in the pep4Δ vps4Δ background; in this strain background the Ath1-HA fusion protein accumulates in its full-length form. Ath1-HA was recovered exclusively in the P13 fraction, as in the wild-type background (Figure 5B and our unpublished results). Next, the P13 fraction was treated with high pH, high salt, or detergent. Ath1-HA was recovered in the supernatant fraction only after detergent treatment (Figure 5B). Pho8, a vacuolar integral membrane protein, served as a positive control and behaved the same as Ath1-HA. These results supported the hypothesis that Ath1 is a transmembrane protein.
Figure 5.
Ath1 is a transmembrane protein with its C terminus in the lumen. (A) Wild-type (WT) and Ath1-HA (YJH1) strains were grown to stationary phase, and the cells were converted to spheroplasts. An aliquot was removed for the total sample, and the remainder was fractionated into supernatant (S13) and pellet (P13) fractions according to Materials and Methods. The Ath1-HA fusion protein was mostly recovered in the P13 fraction. The cytosolic protein Pgk1 serves as a control to monitor spheroplast lysis and separation of the fractions. (B) The Ath1-HA pep4Δ vps4Δ strain (YJH35) was fractionated as above and the P13 fraction was resuspended in either high-salt buffer (1 M KCl), high-pH buffer (0.1 M Na2CO3, pH 11.0), or detergent (1% Triton X-100). After treatment, samples were centrifuged at 13,000 × g for 5 min and separated into supernatant (S) and pellet (P) fractions. Antibodies against HA and Pho8 were used to detect the Ath1-HA fusion protein and Pho8. (C) The P13 fraction from B was resuspended in PS200 buffer in the presence (+) or absence (−) of proteinase K and Triton X-100, as indicated, then resolved by SDS-PAGE, and subjected to Western blot with anti-HA antibody.
As a corollary, to further confirm that the C terminus of Ath1 is in the vacuolar lumen, we carried out a protease protection experiment using the strain in which 3xHA was chromosomally integrated at the C terminus of Ath1 in a pep4Δ vps4Δ background. We have already shown that the C-terminal part of the Ath1-HA fusion protein was not cleaved in the cell (Figure 5A), so we used it to monitor the intact protein. If the C terminus of Ath1 faces the lumen, the HA tail will be protected from exogenously added protease after gentle osmotic lysis. Otherwise, if the C terminus is in the cytosol, it will be digested by external protease, and the fusion protein would not be detected with HA antibody on the Western blot.
Spheroplasts were generated and gently lysed following the methods described in Materials and Methods. The P13 fraction was resuspended in lysis buffer containing 200 mM sorbitol to keep organelles and vesicles intact and treated with proteinase K in the presence or absence of TX-100. The Ath1-HA was protease-insensitive when treated with proteinase K alone, but became sensitive when TX-100 was also present (Figure 5C). These data suggest that the C terminus of Ath1 is in the vacuole lumen. Together, the above results agree with our hypothesis that Ath1 is a type II transmembrane protein with its N terminus in the cytosol and its C terminus in the lumen.
Sorting of Ath1 into the MVB Pathway Is Ubiquitin-independent
Most proteins targeted to the MVB pathway are ubiquitinated on the cytosolic domain, and the attached ubiquitin serves as a sorting signal for internalization into MVB vesicles (Hicke and Riezman, 1996; Roth and Davis, 1996; Katzmann et al., 2001; Reggiori and Pelham, 2001; Chen and Davis, 2002). For example, the yeast vacuolar hydrolase Cps1 is a biosynthetic cargo that is sorted into the MVB pathway for delivery into the vacuole lumen (Felder et al., 1990). Cps1 is synthesized as a type II transmembrane protein in its precursor form, transported through the Golgi network, and then enters into the endosomal system where it is ubiquitinated, recognized by MVB-sorting machinery, and selectively internalized into intralumenal vesicles (Felder et al., 1990). The lysine residue at amino acid 8, which is one of two lysines, K8 and K12, in the cytosolic tail of Cps1 has been found to be the ubiquitination target site. Mutating this lysine to arginine causes Cps1 to mislocalize to the limiting membrane of the vacuole (Katzmann et al., 2001). Phm5 has identical characteristics (Reggiori and Pelham, 2001). Here we have found that Ath1 is also a type II transmembrane protein, and we initially hypothesized that its MVB sorting is ubiquitin-dependent, similar to Cps1 and Phm5. To test our hypothesis, we decided to mutate the lysine residues in the cytosolic domain of Ath1 and examine the localization of the mutant proteins. According to the predicted amino acid sequence, there are three lysines in the Ath1 cytosolic domain, at positions 2, 27, and 37, and we hypothesized that one or more of these residues would be involved in MVB sorting by becoming ubiquitinated. To test this hypothesis, we generated GFP-Ath1 constructs containing mutations of lysine to arginine at one or more of the positions in the cytosolic domain: K2R, K27R, K37R, K27,37R, and K2,27,37R and examined their localization in ath1Δ cells by fluorescence microscopy. There were no differences among the wild-type or mutant proteins, and all of the altered GFP-Ath1 constructs were localized inside the vacuole lumen (Figure 6A). This result suggested that Ath1 could still be targeted into the MVB pathway without ubiquitination.
Figure 6.
Ath1 is sorted into the MVB pathway in an ubiquitin-independent manner. (A) The ath1Δ strain was transformed with pGFPATH1 plasmids encoding Ath1 with the indicated mutations of lysine to arginine in the cytosolic domain. The localization of the chimeric proteins was monitored by fluorescence microscopy. (B) The pGFPATH1, pGFPPHM5, or pGFPSNA3 plasmids were transformed into tul1Δ, bsd2Δ (XGY5), bsd2Δ tul1Δ (YJH38), and fab1Δ mutant strains. Intracellular localization of all GFP-fused proteins was observed by fluorescence microscopy. (C) bsd2Δ cells expressing GFP-Ath1 were grown in SMD-URA media. Cells were collected at early growth phase (OD600 = 0.8–1) and late growth phase (OD600 = 2–3) and subjected to fluorescence microscopy. Differential interference contrast (DIC) images are presented to the right of the GFP images.
To extend this analysis, we examined whether Ath1 transport was blocked in mutants that affect sorting through the MVB pathway other than those involving the ESCRT complexes. Tul1, a transmembrane ubiquitin ligase, is required for ubiquitination of certain vacuolar proteins that utilize the MVB pathway. Fab1 is the phosphatidylinositol(3)phosphate 5-kinase that makes phosphatidylinositol(3,5)P2 (Dove et al., 2002). Proteins that are ubiquitin-dependent for transport, such as Cps1 and Phm5, are mislocalized on the vacuole-limiting membrane in fab1Δ and tul1Δ cells (Figure 6B; Reggiori and Pelham, 2002). Until now, only two proteins have been reported to transit through the MVB pathway in an ubiquitin-independent manner in yeast: Sna3, a possible proton transporter (Reggiori and Pelham, 2001), and Atg15, a putative lipase required for breakdown of autophagic bodies and other intravacuolar vesicles (Epple et al., 2001, 2003; Teter et al., 2001). Here we used Phm5 and Sna3 as controls representing ubiquitin-dependent and -independent targeting proteins, respectively. GFP-Phm5 showed a distribution pattern on the vacuole outer membrane in both tul1Δ and fab1Δ mutants, whereas GFP-Sna3 was detected within the vacuole lumen (Figure 6B). GFP-Ath1 was localized inside the vacuole lumen in both tul1Δ and fab1Δ cells, similar to the result with GFP-Sna3.
To exclude the possibly that localization of Ath1 is dependent on ubiquitination by an ubiquitin ligase other than Tul1, we further examined bsd2Δ and bsd2Δ tul1Δ mutant strains. Bsd2 is an adaptor protein mediating the interaction of cargo with Rsp5, an ubiquitin ligase that is responsible for the attachment of ubiquitin to many proteins including Cps1 and Phm5 (Hettema et al., 2004). In tul1Δ or bsd2Δ cells, Phm5 is at least partially mislocalized to the vacuole membrane, whereas in bsd2Δ tul1Δ cells, almost all of the protein is mislocalized (Hettema et al., 2004). Indeed, GFP-Phm5 was mislocalized to the vacuole membrane in bsd2Δ and bsd2Δ tul1Δ cells (Figure 6B). In contrast, GFP-Sna3 and GFP-Ath1 were both localized to the vacuole lumen.
While examining the bsd2Δ and bsd2Δ tul1Δ mutants, we noticed that GFP-Ath1 was partially retained on the ER membrane when cells were in the early growth phase, in addition to the vacuole lumenal localization (Figure 6C). In the late growth phase the ER staining became less prominent and 80% of the cells displayed primarily a vacuolar localization for GFP-Ath1. In wild-type cells in the early growth phase a small pool of GFP-Ath1 can also be detected on the ER membrane (our unpublished results), but the level is substantially lower than seen in the bsd2Δ and bsd2Δ tul1Δ mutants. Thus, Bsd2 and/or Rsp5 may have some effect on the early transport of Ath1, but not the final stage of vacuolar targeting that involves the MVB pathway. We also investigated the localization of GFP-Ath1 in doa4Δ cells, in which the removal of ubiquitin from cargo proteins is impaired, resulting in a reduction in the free ubiquitin pool. Cps1 and Phm5 are mislocalized to the vacuole outer membrane in doa4Δ cells, but Sna3 is not (Reggiori and Pelham, 2001); however, in the doa4Δ mutants that we tested, we found that even GFP-Sna3 was mislocalized (our unpublished results), probably because of pleiotropic effects in our doa4Δ strain background. Accordingly, we did not pursue the analysis in this mutant. Overall, these results were consistent with our analysis of Ath1 containing mutated lysine residues and further suggest that Ath1 can be sorted into the MVB pathway independent of ubiquitination.
Ath1 Is Ubiquitinated on Its Cytosolic Tail
Although ubiquitination of the cytosolic lysine residues was not required for correct localization of Ath1, we decided to examine whether the protein undergoes ubiquitination. Cell lysates from wild-type and Ath1-HA strains were subjected to immunoprecipitation with anti-HA antibody. The precipitated protein was then examined by Western blot using antibodies to either HA or ubiquitin. The Ath1-HA protein band was detected in the HA-tagged strain, but not in wild-type cells (Figure 7A), which suggested that we were able to specifically recognize the tagged protein. The Ath1-HA protein was also detected by anti-ubiquitin antibody (Figure 7A). The band corresponding to the ubiquitinated species was very faint. This may represent transient ubiquitination of Ath1, with the major pool of the protein existing in the de-ubiquitinated form.
Figure 7.
Ath1 is ubiquitinated on its cytosolic domain. (A) Wild-type (WT) and Ath1-HA (YJH1) cells were immunoprecipitated with anti-HA antibody as described in Materials and Methods. The immunoprecipitate was subjected to SDS-PAGE and immunoblotted with anti-HA or anti-Ub antibody. The precipitated Ath1-HA protein was detected with anti-Ub antibody, indicated as Ub-Ath1-HA. (B) doa4Δ (MHY623) cells were transformed with plasmids expressing HA-Ub alone, GFP-Ath1 alone or together with HA-Ub, or GFPAth1K2,27,37R (GFP-Ath1KR) alone or together with HA-Ub. Cell lysates were immunoprecipitated with anti-HA antibody and then immunoblotted with anti-GFP and anti-Cps1 antibody or antiserum. The asterisk indicates a nonspecific band.
To verify this result and further test whether ubiquitination occurs on the lysine residues of the Ath1 cytosolic tail, we carried out immunoprecipitation experiments in cells expressing ubiquitin tagged with HA (HA-Ub; Hochstrasser et al., 1991). We used the doa4Δ background to reduce de-ubiquitination and potentially increase the pool of ubiquitinated Ath1. Protein extracts from cells expressing combinations of HA-Ub, GFP-Ath1, and/or GFP-Ath1K2,27,37R were immunoprecipitated with anti-HA antibody. The total lysate and precipitate were then subjected to immunoblotting with anti-GFP antibody. GFP-Ath1 or GFP-Ath1K2,27,37R were only detected in lysates from the cells that expressed the corresponding plasmids (Figure 7B). When cells carried plasmids expressing both HA-Ub and GFP-Ath1, the GFP-Ath1 was coprecipitated with HA-Ub and was detected on the Western blot with GFP antibody (Figure 7B). Under these conditions, the level of ubiquitinated GFP-Ath1 was substantially higher than seen in the wild-type background (Figure 7A). In contrast to the results with GFP-Ath1, GFP-Ath1K2,27,37R, which lacks the lysines in the cytosolic tail, was not coprecipitated with HA-Ub and subsequently could not be detected with anti-GFP. As a positive control, the ubiquitinated form of Cps1 was detected in samples from all three strains that expressed HA-Ub, but not from strains that did not harbor this plasmid (Figure 7B). These results suggested that Ath1 is normally ubiquitinated and that mutation of the lysine residues in its cytosolic tail prevented this modification.
The Transmembrane Domain of Ath1 Contains an MVB Sorting Signal
Currently only Sna3 and Atg15 have been shown to transport into the vacuole via the MVB pathway in an ubiquitin-independent manner, but it is still unknown what signals direct them into the MVB pathway. Recent studies showed that Sna3 is also ubiquitinated; however, without ubiquitination it still can utilize an alternative mechanism, which involves the function of Rsp5, to be sorted into the MVB pathway (McNatt et al., 2007; Oestreich et al., 2007). Here we showed that the acid trehalase, Ath1, had the same transport behavior as Sna3. Ath1 is ubiquitinated, but it can be also targeted into the MVB pathway in an ubiquitin-independent manner. To understand the ubiquitin-independent sorting mechanism of Ath1, we decided to examine which region contains the sorting signal. We generated three constructs, pGFPAth1ΔC, pGFPAth1ΔN, and pGFPAth1TM, which express Ath1 lacking the lumenal C-terminal domain, or the cytoplasmic N-terminal tail or contain only the transmembrane domain of Ath1, respectively. Both Ath1ΔC and Ath1ΔN were still mostly delivered into the vacuole lumen, suggesting that neither the cytosolic tail nor lumenal part of the protein are essential for diversion into the MVB pathway (Figure 8A). Similarly, the transmembrane domain of Ath1 expressed alone was readily transported into the vacuole. This result suggested that the transmembrane domain of Ath1 contains a sorting signal that is sufficient for sorting into the MVB pathway.
Figure 8.
The transmembrane domain of Ath1 contains a sufficient signal for sorting into the MVB pathway. (A) The pGFPATH1ΔC, pGFPATH1ΔN, and pGFPATH1TM constructs (shown schematically on the right) were transformed into ath1Δ cells separately to express GFP-tagged Ath1 containing the N terminus and transmembrane domains, the C terminus and transmembrane domains, or only the transmembrane domain, respectively. All of the altered proteins were still localized within the vacuole lumen. (B) The fab1Δ, tul1Δ, bsd2Δ tul1Δ, and wild-type (WT) strains expressing either wild-type GFP-Phm5 or GFP-Phm5/Ath1TM, in which the original transmembrane domain of Phm5 was replaced with the transmembrane domain of Ath1, were examined by fluorescence microscopy. Differential interference contrast (DIC) images are presented to the right of the GFP images in A and B. (C) Wild-type cells transformed with plasmids expressing GFP-Ath1ΔN, GFP-Ath1polarmut (in which four polar amino acid residues in the transmembrane domain had been replaced with hydrophobic residues), or GFP-Ath1ΔNpolarmut mutant proteins were monitored by fluorescence microscopy. Images from 500 randomly selected cells containing a GFP signal from each strain were analyzed, and cells that showed a weak or partial vacuolar membrane fluorescent signal were counted. The percentage of cells displaying vacuolar membrane fluorescence for each construct is shown beneath the corresponding image. The white arrowhead points to the ER.
Further analysis by swapping the Ath1 transmembrane domain for that of the ubiquitin-dependent MVB cargo protein, Phm5, confirmed this result. As illustrated in Figure 8B, Phm5 was delivered to the vacuole lumen in the wild-type strain, but could not be efficiently targeted to the interior of the vacuole in tul1Δ, bsd2Δ tul1Δ, or fab1Δ mutants and was instead largely mislocalized to the vacuole outer membrane. In contrast, most of the chimeric Phm5/Ath1TM protein was visible in the vacuole lumen when the Ath1 transmembrane domain replaced that of Phm5. These results implied that the transmembrane domain of Ath1 contains an efficient sorting signal that directed the protein into the multivesicular body vesicles, which were later delivered into the vacuole lumen.
Although the Ath1 transmembrane domain was quite efficient at directing GFP into the vacuole lumen, we found that a small percentage of the cells expressing Ath1ΔN, ∼12%, displayed a small pool of the protein on the vacuole membrane. This observation suggests that the transmembrane domain may not be the only region containing the MVB-sorting signal. An additional signal located in the cytosolic tail of Ath1, possibly dependent upon ubiquitination, may provide an alternative mechanism for Ath1 targeting. Our finding that the Ath1 transmembrane domain itself can be directly or indirectly recognized by ESCRT machinery is novel. To understand the details of the recognition sites within the transmembrane region, we further carried out mutagenesis of the 23 amino acid membrane-spanning region. There are four polar residues in the transmembrane domain: asparagine at position 49, serine 50, threonine 65, and tyrosine 68. These residues are not typical for a highly hydrophobic integral membrane domain and are not in the conserved core region that serves as the signal peptide recognized by the ER translocon; therefore we hypothesized that these polar residues might be involved in MVB sorting.
Substitution of all four polar amino acids with hydrophobic residues resulted in the majority of GFP-Ath1polarmut being localized to the vacuole lumen (Figure 8C); however, in ∼5% of the cells, a faint vacuole outer membrane staining was detected, similar to the low level seen with the GFP-Ath1ΔN protein. Considering that the N-terminal region may contain a redundant (ubiquitin-dependent) targeting signal, we further made the polar residue mutations in the N-terminal truncated form of Ath1, GFP-Ath1ΔN, and monitored the cellular localization of the resulting GFP-Ath1ΔNpolarmut construct. We found that in ∼75% of the cells, the mutant protein was partially mislocalized to the vacuolar membrane (Figure 8C). In addition, the extent of the missorting defect was more severe than seen with either the GFP-ATh1ΔN or GFP-Ath1polarmut mutations alone. These results suggest that the polar residues in the transmembrane domain are involved in the recognition by the MVB-sorting machinery.
DISCUSSION
Glycosylation and Localization of Acid Trehalase
Keller et al. (1982) first demonstrated that an active trehalase zymogen resides in the vacuole. Later, this enzyme was shown to give a maximal activity at pH 4.5 and thus was referred to as acid trehalase (Mittenbuhler and Holzer, 1988). Vacuolar glycoproteins are usually modified by uniform N-linked oligosaccharides at specific sites and the resulting glycan chains have the same molecular mass. In the case of most secreted proteins, individual molecules of the same protein often undergo differential levels of glycosylation, which generates a mixture of differently sized protein species that migrate as a smear during SDS-PAGE. This type of heterogeneous glycosylation has not been seen for any identified vacuolar hydrolases. Here we examined a GFP-Ath1 fusion protein expressed under the control of its endogenous promoter and a strong heterologous promoter and found that the chimera migrated as a sharp band rather than a smear when resolved by SDS-PAGE (Figure 1). Consistent with this finding, endogenous Ath1 chromosomally tagged with HA was also detected as only one sharp band by Western blot, confirming that the protein only undergoes limited glycosylation (Figure 5). Endoglycosidase H treatment resulted in a molecular mass shift of the protein, and an apparent reduction in size of ∼60 kDa (Figure 4). These results suggested that Ath1 is a glycoprotein with only limited glycosyl modification. According to its amino acid sequence, there are 26 potential N-linked glycosylation sites. With limited glycosylation, each N-linked oligosaccharide unit is ∼2.5 kDa. It follows that if all 26 sites are modified, the glycosylated protein will be 65 kDa larger in molecular mass than the unglycosylated form, which fits with our results and is consistent with Ath1 being a vacuole resident hydrolase.
Controversy concerning the vacuolar localization of Ath1 resulted from a recent study by Jules et al. (2004). They demonstrated that 90% of the total cellular acid trehalase activity is extracellular, in the cell wall or on the plasma membrane. They suggested that the reason why no activity was detected when incubating intact yeast cells with trehalose was due to the rapid uptake into the cells of the glucose generated by trehalase-dependent cleavage of trehalose. When sodium fluoride, which impairs the transport of glucose into cells, was used to treat intact yeast cells, 90% of the acid trehalase activity was recovered in the extracellular space. By contrast, both our data from fluorescence microscopy and enzyme assay with purified vacuoles showed a clear localization of Ath1 in the vacuole lumen. We also generated spheroplasts by removing the cell wall and collected the extracellular fraction. If the majority of Ath1 is located in the cell wall, this fraction should demonstrate the most enzyme activity; however, we did not detect any activity in this fraction and instead, most of the activity was found in the spheroplast lysates (our unpublished results). To further investigate whether or not Ath1 is on the plasma membrane, we looked at the protease sensitivity of the C-terminal HA-tagged Ath1. We found that the C terminus of Ath1 faces the vacuolar lumen, and was protected from exogenous proteinase (Figure 5). Accordingly, it is not possible that this protein is on the plasma membrane with a topology that would provide extracellular activity. Moreover, results from Jules et al. (2004) may have been misleading due to the treatment with sodium fluoride. Sodium fluoride is toxic to yeast because it inhibits glucose phosphorylation, which is involved in the active transport and retention of glucose in the cell. Once glucose phosphorylation is impaired, intracellular free glucose can be released from the cell, moving down the glucose concentration gradient.
Transport of Acid Trehalase
Ath1 transits through the MVB pathway and is delivered to the vacuole. To date there has been no solid clue to show how Ath1 is delivered to this organelle. One model suggests that after it is translocated into the ER and reaches the Golgi complex, the acid trehalase is delivered to the periplasmic space where it binds extracellular trehalose and moves to the vacuole by endocytosis. In another model, Ath1 and trehalose are sorted into the vacuole separately. Here we present the first study illustrating the biosynthetic pathway of Ath1 and we show that its vacuolar transport is through the MVB pathway. Multivesicular bodies are the convergence point between the delivery route from the TGN to the lysosomes/vacuoles and endocytosis (Katzmann et al., 2002; Gruenberg and Stenmark, 2004; Babst, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006). However, our analysis of the GFP-Ath1 chimera in the end3Δ mutant demonstrates that Ath1 reaches its final localization from the TGN via endosomes without transiting to the plasma membrane (Figure 3). This finding indicates that Ath1 does not bind its substrate on the plasma membrane and thus suggests that the extracellular trehalose also moves to the vacuole through either endocytosis or another transport pathway. Thus, identification of the trafficking route of Ath1 further helps us understand the way it functions and the process of trehalose metabolism.
A New Signal for Protein Sorting into MVB Internal Vesicles
Most proteins trafficking through the MVB pathway require ubiquitination as a sorting signal (Babst, 2005; Hurley and Emr, 2006; Slagsvold et al., 2006). In rare cases, however, some proteins are internalized into multivesicular bodies without ubiquitination. Sna3 and Atg15 in yeast and the δ-opioid receptor and Pmel17 in mammalian cells are the only proteins reported to transit in this manner (Reggiori and Pelham, 2001; Epple et al., 2003; Hislop et al., 2004; Theos et al., 2006). Furthermore, it remains questionable whether sorting of Atg15 is indeed ubiquitin-independent, because it was shown to be mislocalized to the vacuolar membrane in a fab1Δ mutant, but not affected in a tul1Δ mutant (Epple et al., 2001, 2003). Tul1 has redundant activity with the Rsp5-Bsd2 complex, and therefore a possible explanation of the published result could be that Atg15 is a better substrate for the Rsp5 ligase than for Tul1 (Hettema et al., 2004).
Here we found that Ath1 is a bona fide member of this ubiquitin-independent group. The mechanism of targeting these proteins into the MVB pathway is unknown. Sna3 and the δ-opioid receptor are multispan transmembrane proteins, and the identification of their localization signal with sequential truncations is particularly difficult because the transmembrane domains have crucial folding and structural roles. Ath1, on the other hand, has a single transmembrane segment, which permits the use of a deletion and/or mutational analysis. In this study we have found that the domain of Ath1 spanning the lipid bilayer contains sufficient signaling information to target the protein into the MVB pathway (Figure 7A). Importantly, transplantation of this domain into Phm5, a protein with a topological organization similar to Ath1 but that requires ubiquitination to enter the MVB internal vesicles (Reggiori and Pelham, 2001), allows this phosphatase to enter the same structures in an ubiquitin-independent manner (Figure 7B). Transmembrane domains have already been implicated as a motif for protein sorting in essentially all the compartments of the secretory pathway and of the endosomal system, with polar and charged residues playing a pivotal role in the localization process (Bonifacino et al., 1990; Munro, 1995; Sato et al., 1996; Rayner and Pelham, 1997; Letourneur and Cosson, 1998; Lewis et al., 2000; Reggiori et al., 2000; Reggiori and Pelham, 2002). In particular, it has been shown that introduction of polar residues into the transmembrane domain of the endosomal SNARE, Pep12, directed it into MVB vesicles that were transported into the vacuole (Reggiori et al., 2000). In this specific case, however, Pep12 is recognized as an aberrant protein and ubiquitination by Tul1 targets it for destruction (Reggiori and Pelham, 2002; Hettema et al., 2004). Here we shown that the polar residues in the Ath1 transmembrane segment play a crucial role in delivering this protein to the vacuole through the MVB pathway.
A first major implication of our findings is that eukaryotic cells possess at least two different systems to deliver integral membrane proteins into the lysosome/vacuole interior, allowing them to have the flexibility necessary to correctly sort the numerous biosynthetic and endocytosed cargoes, including those without cytoplasmic lysines or without accessible ones. These two sorting systems are not mutually exclusive and probably coexist in certain proteins to provide more efficient localization. For example, Sna3 is ubiquitinated and that probably facilitates its sorting into multivesicular bodies (Peng et al., 2003). Along these lines, recent studies show that Sna3 can utilize both ubiquitin-dependent and -independent mechanisms for its MVB targeting. Sna3 ubiquitination is dependent on Rsp5, but the function of Rsp5 is also critical for the MVB sorting of Sna3 independent of cargo ubiquitination (McNatt et al., 2007; Oestreich et al., 2007). Here we found that Ath1 has certain properties that are similar to Sna3. In particular, Ath1 can be sorted into the MVB pathway through both ubiquitin-dependent and -independent routes; however, it appears to utilize a mechanism that is independent of Bsd2-Rsp5 (Figure 8).
A second important implication of our study regards the mechanism of MVB biogenesis. The currently accepted model is that the association of the Vps27/Hrs-Hse1/STAM (ESCRT-0) complex to the endosomal membrane via its simultaneous binding to phosphatidylinositol(3)phosphate and ubiquitinated cargoes induces the recruitment of ESCRT-I (Babst, 2005; Slagsvold et al., 2006). This event triggers a series of sequential reactions, including the recruitment of the two other ESCRT complexes, which ultimately leads to the formation of MVB internal vesicles. Our finding evokes the possibility of an alternative mechanism dependent on the transmembrane domain of certain MVB cargo proteins for triggering this process. This idea is supported by the fact that in mutants such as doa4Δ and bsd2Δ tul1Δ, MVB internal vesicles are generated in the absence of ubiquitinated cargoes (Reggiori and Pelham, 2001, 2002). The mechanism by which transmembrane domains could induce MVB biogenesis through interaction with the cytosolic Vps27/Hrs-Hse1/ STAM complex and ESCRT-I factors is not known; however, our results suggest that the polar residues play a role in the signal recognition. Additional analysis of Ath1 transport may provide further information about the mechanism of protein sorting into the MVB pathway.
ACKNOWLEDGMENTS
The authors thank Dr. Amy Chang (University of Michigan) for providing strain XGY5. This work was supported by the National Institutes of Health Public Health Service Grant GM53396 to D.J.K.
Abbreviations used:
- Ath1
acid trehalase
- ER
endoplasmic reticulum
- MVB
multivesicular body
- Nth1
neutral trehalase
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-11-0995) on May 2, 2007.
REFERENCES
- Alizadeh P., Klionsky D. J. Purification and biochemical characterization of the ATH1 gene product, vacuolar acid trehalase, from Saccharomyces cerevisiae. FEBS Lett. 1996;391:273–278. doi: 10.1016/0014-5793(96)00751-x. [DOI] [PubMed] [Google Scholar]
- Avaro S., Belgareh-Touze N., Sibella-Arguelles C., Volland C., Haguenauer-Tsapis R. Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast. 2002;19:351–371. doi: 10.1002/yea.838. [DOI] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Cosson P., Klausner R. D. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Chen L., Davis N. G. Ubiquitin-independent entry into the yeast recycling pathway. Traffic. 2002;3:110–123. doi: 10.1034/j.1600-0854.2002.030204.x. [DOI] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe L. M., Mouradian R., Crowe J. H., Jackson S. A., Womersley C. Effects of carbohydrates on membrane stability at low water activities. Biochim. Biophys. Acta. 1984;769:141–150. doi: 10.1016/0005-2736(84)90017-8. [DOI] [PubMed] [Google Scholar]
- Dove S. K., McEwen R. K., Mayes A., Hughes D. C., Beggs J. D., Michell R. H. Vac14 controls PtdIns(3,5)P2 synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 2002;12:885–893. doi: 10.1016/s0960-9822(02)00891-6. [DOI] [PubMed] [Google Scholar]
- Elbein A. D. The metabolism of α-α-trehalose. Adv. Carbohydr. Chem. Biochem. 1974;30:227–256. doi: 10.1016/s0065-2318(08)60266-8. [DOI] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Epple U. D., Eskelinen E.-L., Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Epple U. D., Suriapranata I., Eskelinen E.-L., Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder S., Miller K., Moehren G., Ullrich A., Schlessinger J., Hopkins C. R. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 1990;61:623–634. doi: 10.1016/0092-8674(90)90474-s. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Haas A. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 1995;17:283–294. [Google Scholar]
- Harris S. D., Cotter D. A. Vacuolar (lysosomal) trehalase of Saccharomyces cerevisiae. Curr. Microbiol. 1987;15:247–249. [Google Scholar]
- Harris S. D., Cotter D. A. Transport of yeast vacuolar trehalase to the vacuole. Can. J. Microbiol. 1988;34:835–838. doi: 10.1139/m88-143. [DOI] [PubMed] [Google Scholar]
- Hettema E. H., Valdez-Taubas J., Pelham H.R.B. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–1288. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hislop J. N., Marley A., Von Zastrow M. Role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J. Biol. Chem. 2004;279:22522–22531. doi: 10.1074/jbc.M311062200. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E., Davidson A. R., Kaiser C. A. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M., Cai Z. L., Ho S. N., Pease L. R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins M. U., Klionsky D. J. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Bankaitis V. A., Emr S. D. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987;48:875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Jules M., Guillou V., Francois J., Parrou J. L. Two distinct pathways for trehalose assimilation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2004;70:2771–2778. doi: 10.1128/AEM.70.5.2771-2778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Keller F., Schellenberg M., Wiemken A. Localization of trehalase in vacuoles and of trehalose in the cytosol of yeast (Saccharomyces cerevisiae) Arch Microbiol. 1982;131:298–301. doi: 10.1007/BF00411175. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cueva R., Yaver D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M., Muller H., Holzer H. Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J. Biol. Chem. 1993;268:4766–4774. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Letourneur F., Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J. Biol. Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]
- Lewis M. J., Nichols B. J., Prescianotto-Baschong C., Riezman H., Pelham H.R.B. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marcusson E. G., Horazdovsky B. F., Cereghino J. L., Gharakhanian E., Emr S. D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- McNatt M. W., McKittrick I., West M., Odorizzi G. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol. Biol. Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenbuhler K., Holzer H. Purification and characterization of acid trehalase from the yeast suc2 mutant. J. Biol. Chem. 1988;263:8537–8543. [PubMed] [Google Scholar]
- Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S., Holzer H. Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:197–237. doi: 10.1016/s0079-6603(08)60037-9. [DOI] [PubMed] [Google Scholar]
- Nwaka S., Mechler B., Holzer H. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett. 1996;386:235–238. doi: 10.1016/0014-5793(96)00450-4. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S. D. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Oestreich A. J., Aboian M., Lee J., Azmi I., Payne J., Issaka R., Davies B. A., Katzmann D. J. Characterization of multiple multivesicular body sorting determinants within Sna3, A role for the ubiquitin ligase Rsp5. Mol. Biol. Cell. 2007;18:707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opheim D. J. α-D-Mannosidase of Saccharomyces cerevisiae. Characterization and modulation of activity. Biochim. Biophys. Acta. 1978;524:121–130. doi: 10.1016/0005-2744(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Papa F. R., Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Raths S., Rohrer J., Crausaz F., Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner J. C., Pelham H.R.B. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 1997;16:1832–1841. doi: 10.1093/emboj/16.8.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Black M. W., Pelham H.R.B. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 2000;11:3737–3749. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R.B. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R.B. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 2002;4:117–123. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- Rieder S. E., Emr S. D. Somerset, NJ: John Wiley & Sons; 2000. Current Protocols in Cell Biology; pp. 3.7.1–3.7.11. [Google Scholar]
- Roth A. F., Davis N. G. Ubiquitination of the yeast a-factor receptor. J. Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell M.R.G., Nickerson D. P., Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 2006;18:422–428. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Sato M., Sato K., Nakano A. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J. Cell Biol. 1996;134:279–293. doi: 10.1083/jcb.134.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Teter S. A., Eggerton K. P., Scott S. V., Kim J., Fischer A. M., Klionsky D. J. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G., Marks M. S. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev. Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M. Cyclic-AMP content and trehalase activation in vegetative cells and ascospores of yeast. Arch Microbiol. 1984;138:64–67. doi: 10.1007/BF00425409. [DOI] [PubMed] [Google Scholar]
- Van Dijck P., Colavizza D., Smet P., Thevelein J. M. Differential importance of trehalose in stress resistance in fermenting and nonfermenting Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 1995;61:109–115. doi: 10.1128/aem.61.1.109-115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T. A., Emr S. D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn R., Stevenson B. J., Schroder-Kohne S., Zanolari B., Riezman H., Munn A. L. End13p/Vps4p is required for efficient transport from early to late endosomes in Saccharomyces cerevisiae. J. Cell Sci. 2001;114:1935–1947. doi: 10.1242/jcs.114.10.1935. [DOI] [PubMed] [Google Scholar]