Figure 3.
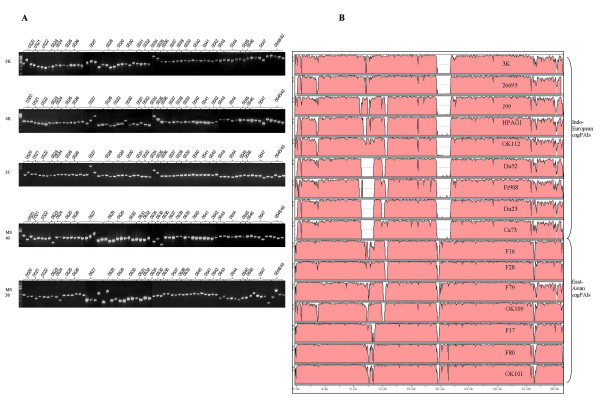
Comparative genomic analysis of the cagPAIs from Indian isolates. A) PCR based analysis of the complete cagPAI of 5 representative hpEurope Isolates: 3K, 4K, 3C, MS40 and MS38 from India. Overlapping PCR primers amplified the whole cagPAI indicating the intactness of the PAI in these isolates. B) Global pair-wise alignments of whole cagPAI sequences of different H. pylori isolates were generated by VISTA using default parameters [47]. The OK129 genome was taken as the base sequence (not shown) and rest of the sequences were aligned against it. The X-axis denotes length of the sequence under consideration and the Y-axis conveys homology in % with the base genome sequence). The Indian hpEurope isolate, 3K was aligned with other whole cagPAI sequences from GenBank along with the cagPAIs of HP 26695, HPJ99 and HPAG1. The accession numbers for the public domain sequences of the cagPAIs from Europe [9] and Japan [49] that we used in our analyses, were as follows – Ca73 (AY330638 and AY330639), Du23:2 (AY330643 and AY330644), Du52:2 (AY330640, AY330641 and AY330642), F80 (AB120421), OK112 (AB120425), F16 (AB120416), F17 (AB120417), F28 (AB120418), F79 (AB120420), OK101 (AB120422), OK109 (AB120424). Sequence of the French isolate, Fr 908 was determined in this study (EF195721). While the cagPAI sequence of the Indian isolate 3K (hpEurope) was found to be genetically highly similar to and aligning closely with the 26695 sequence, it also revealed significant sequence similarities with other isolates of European origins (that harbor Western type of cag EPIYA sequences) such as HPAG1, OK112, Du52, Du23, Ca73, J99 and Fr908. It was however largely unrelated to the East Asian like isolates (mainly harboring Asian type cag EPIYA sequences) such as F16, F28, F79, OK109, F17, OK101 and F80.
