Figure 4.
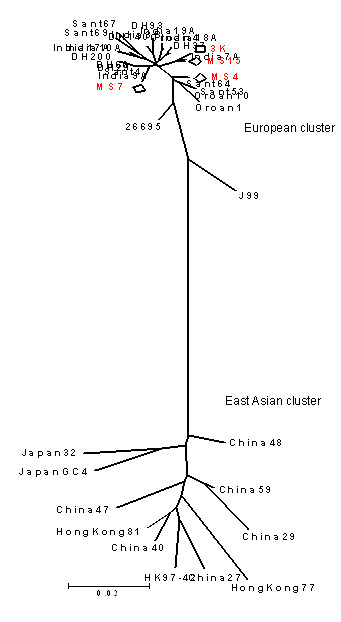
Phylogenetic tree based on the 5' end sequence of the cagA (an informative 219 bp segment of cagA was used to align sequences from unrelated isolates) suggests possible common origins for isolates from ethnic Indians and the tribal. Representative Indian genotypes (3K, MS4, Ms7 and MS15) based on this 219 bp sequence clustered tightly with previously determined genotypes of strains obtained from ethnic Bengalis [India3B (AF202219), India7A (AF202220), India9A (AF202221), India10A (AF202222), India17A (AF202223), India18A (AF202224), India19A (AF202225), DH140 (AY169293), DH200 (AY169294), DH29 (AY169295), DH37 (AY169296), DH60 (AY169297), DH93 (AY169298)] and Santhal and Oraon tribals [Sant4 (AY162446), Sant53 (AY162447), Sant64 (AY162448), Sant67 (AY162449), Sant69 (AY162450), Oraon1 (AY162451), Oraon10 (AY162452), Oraon4 (AY162453)] [20]. All the East Asian strains [China27 (AJ252979), China29 (AJ252980), China40 (AJ252982), China48 (AJ252983), China47 (AJ252985), China59 (AJ252986), Hongkong77 (AF198485), Hongkong81 (AF198486), Hongkong97-42 (AF239733), Japan GC4 (AF198484), Japan32 (AJ239726)], however, clustered together and formed a separate cluster.
