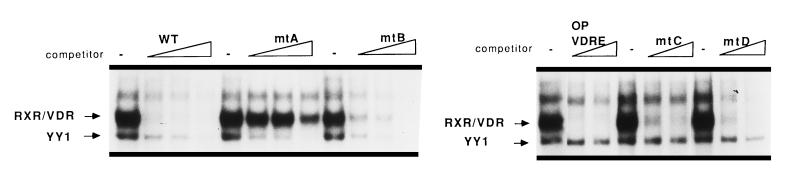
Figure 3.
YY1 recognizes the proximal 5′-CAT-3′ core motif. EMSAs were carried out to determine the location of the YY1 binding motif within the rOC-VDRE using a panel of nucleotide substitution mutations: mtA, mtB, mtC, and mtD (Fig. 1A). These mutants were designed to abrogate either the distal or proximal YY1 binding motif, or both. Binding reactions contain the rOC-VDRE as probe and nuclear proteins from vitamin D (10−8 M, 24 hr)-treated ROS 17/2.8 cells. All competition assays were performed using a 10-, 25-, or 50-fold molar excess of competitor oligonucleotides (as indicated above the lanes), with the exception of the right portion with 25- or 50-fold molar excess, respectively.