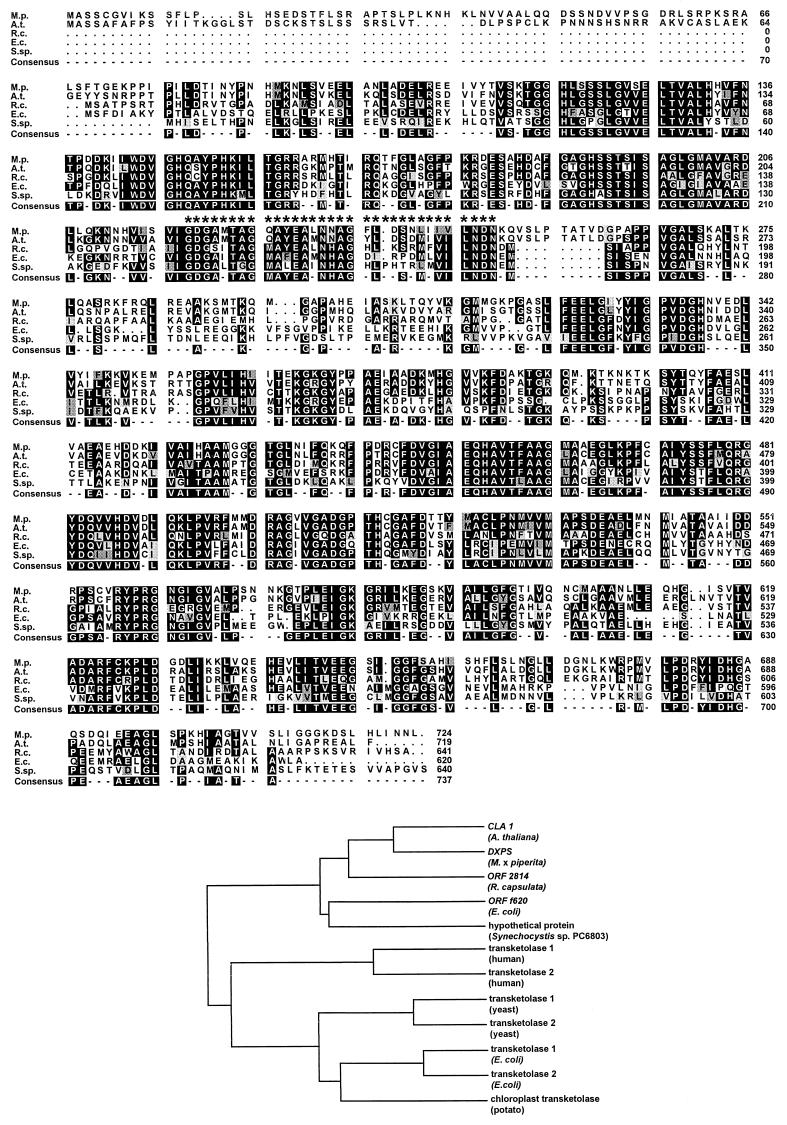
Figure 4.
Deduced amino acid sequence comparison (upper panel) of DXPS from peppermint (M.p.), CLA1 from A. thaliana (A.t.), ORF 2814 from R. capsulata (R.c.), ORF f620 from E. coli (E.c.), and a protein of unknown function from Synechocystis sp. strain PC6803 (S.sp.). Identical residues are shaded in black, residues of high similarity are indicated by gray shading, and residues of lower similarity are displayed by a pale shade. Asterisks indicate the position of the putative TPP-binding motif. Sequence comparison and clustering relationship analysis (lower panel) were carried out by using GCG version 9.0 of the University of Wisconsin Genetics Computer Group Package (1997). The following transketolase sequences are included: DXPS (M. × piperita, accession number AF019383); CLA1 (A. thaliana, U27099); ORF 2814 (R. capsulata, P26242); ORF f620 (E. coli, U82664); a protein of unknown function (Synechocystis sp. PC6803, D90903); transketolase 1 (human, A45050; yeast, P23254; and E. coli, P27302); transketolase 2 (human, P51854; yeast, P33315; E. coli, P33570); and a plastidial transketolase from potato (Z50099).