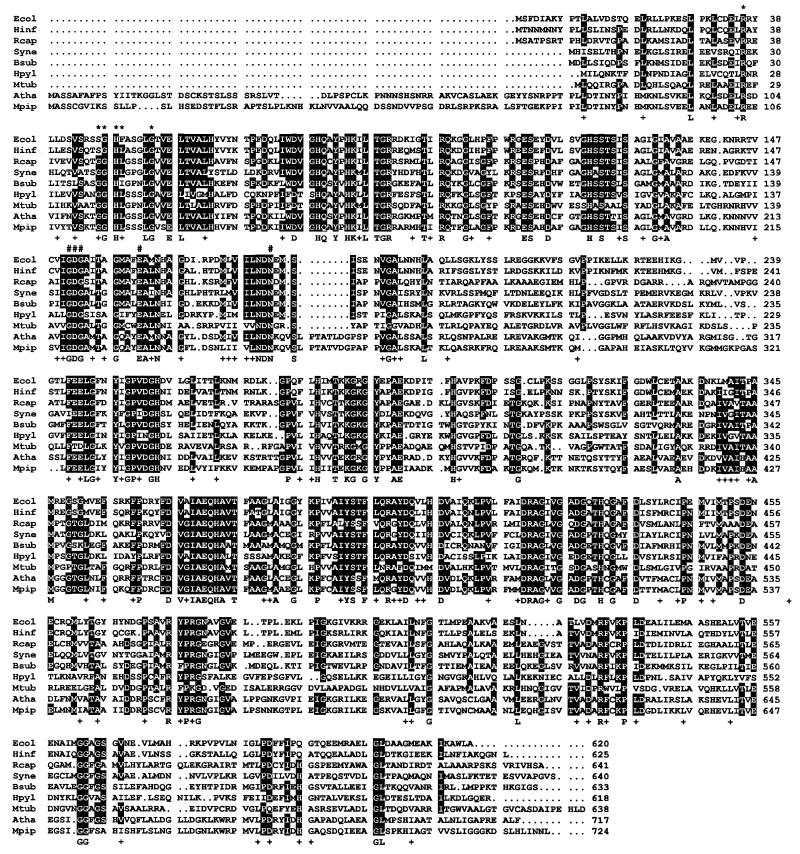
Figure 3.
Amino acid sequence alignment of d-1-deoxyxylulose-5-phosphate synthase from different organisms. Dots indicate the absence of particular amino acid residues. Numbers indicate the position of amino acid residues in the sequences. White-on-black letters denote amino acid residues common to at least six polypeptides. A consensus sequence denoting identical and conserved amino acid residues in all polypeptides is shown below the alignment. Conserved residues are indicated with the symbol +. Amino acid residues putatively involved in the binding of thiamin diphosphate are indicated with the symbol #. The conserved motif containing the histidine residue putatively involved in proton transfer during catalysis is indicated with ∗. Ecol, E. coli (GenBank AF035440); Hinf, H. influenzae (Swiss-Prot P45205); Rcap, R. capsulatus (Swiss-Prot P26242); Syne, Synechocystis sp. (PCC6803) (GenBank D90903); Bsub, B. subtilis (Swiss-Prot P54523); Hpyl, H. pylori (GenBank AE000552); Mtub, M. tuberculosis (GenBank Z96072); Atha, A. thaliana (GenBank U27099); Mpip, Mentha × piperita [GenBank AF019383; Lange et al. (40)].