Figure 2. Mutation of Aspartate 784 to Glycine in A135 Impairs Pol I Elongation Rate.
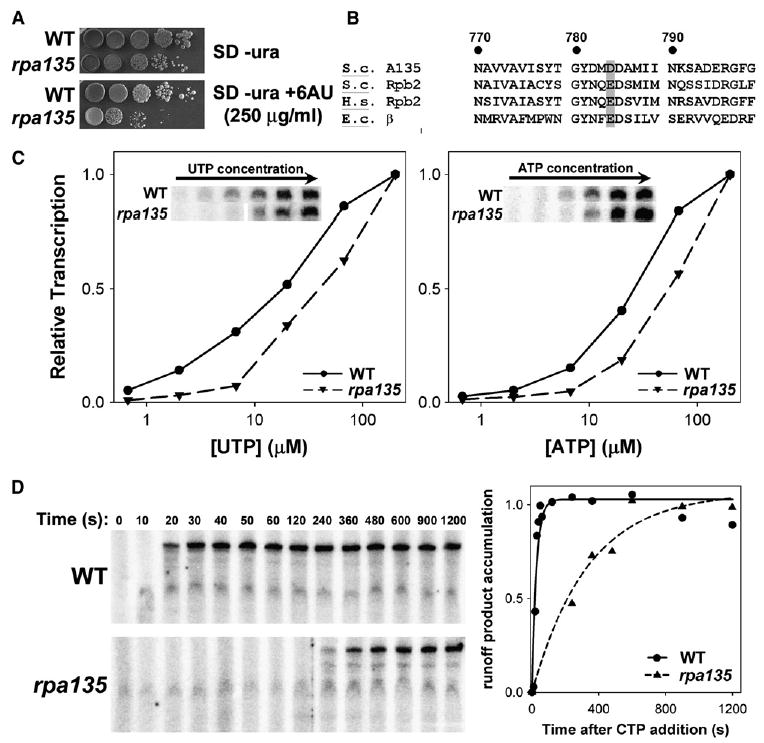
(A) Ten-fold dilutions of yeast cells with WT A135 (NOY388) and rpa135(D784G) (NOY2172) carrying pRS316 were plated on SD −Ura medium and SD −Ura +250 μg/ml 6AU.
(B) Protein sequences flanking D784 of S. cerevisiae (S.c.) A135 aligned with S.c. Rpb2, Homo sapiens (H.s.) Rpb2, and Escherichia coli (E.c.) β. Numbering is relative to S.c. A135, and conserved acidic residues equivalent to D784 are shaded in gray.
(C) Multiround in vitro transcription assays varying the concentration of UTP (left) or ATP (right) using purified WT or rpa135(D784G) Pol I. Runoff product accumulation was normalized to the value at the highest NTP concentration (the values were as follows: for UTP, WT 6983 and rpa135 6893; for ATP, WT 5328 and rpa135 8254 in arbitrary units) and plotted versus the varied NTP concentration. Raw data are inset in each plot.
(D) Elongation rate assay measuring runoff product (763 nt) accumulation versus time after release of elongation complexes arrested at +56 (relative to transcription start site) using WT and rpa135(D784G) mutant Pol I. Raw data are shown on the left, with quantification of the amount of runoff product normalized to maximum and plotted versus time on the right.
