Figure 3. rRNA Accumulation and Ribosome Assembly Are Impaired in the rpa135(D784G) Mutant Compared to WT.
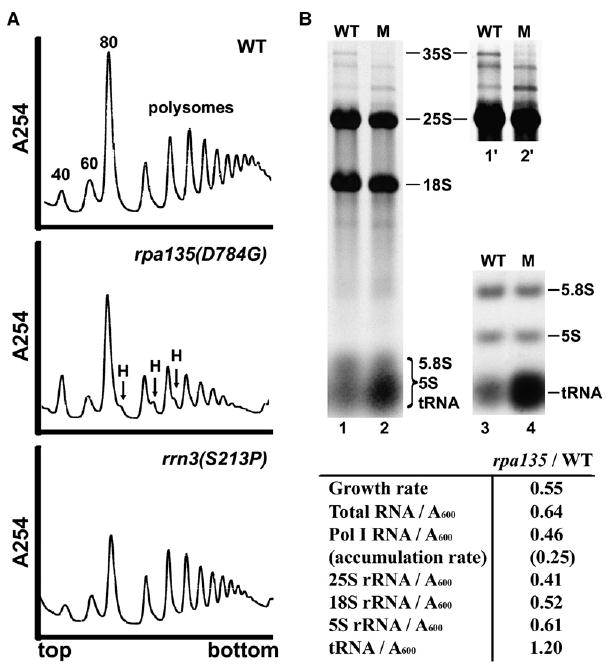
(A) Sucrose gradient analysis of ribosome profiles in WT (NOY388), rpa135(D784G) (NOY2172), and rrn3(S213P) (NOY1075). Cells were grown in YEPD at 30°C, and extracts containing approximately equal amounts of RNA were analyzed by sucrose gradient centrifugation. Positions of 40S, 60S, 80S, polysomes, and halfmers (H) are indicated.
(B) RNA was purified from WT and rpa135(D784G) (lanes labeled “M”) cells that were grown in SD −Ura containing [14C]uracil (1 μCi/ml; 5 μg/ml). RNA containing equal amounts of 14C radioactivity was analyzed on formaldehyde-agarose gels (lanes 1 and 2) or agarose:acrylamide composite gels (lanes 3 and 4; only a lower part shown). Lanes 1′ and 2′ are 3-fold longer exposures of the upper part of lanes 1 and 2, for visualization of low abundance RNAs. The locations of various RNA species are indicated. RNA species were excised from the gels after autoradiography, and amounts of individual species were quantified. The amounts of individual RNA species in the mutant cells were normalized to WT and are summarized below.
