Figure 5. The 5′-S1 Fragment Is Polyadenylated.
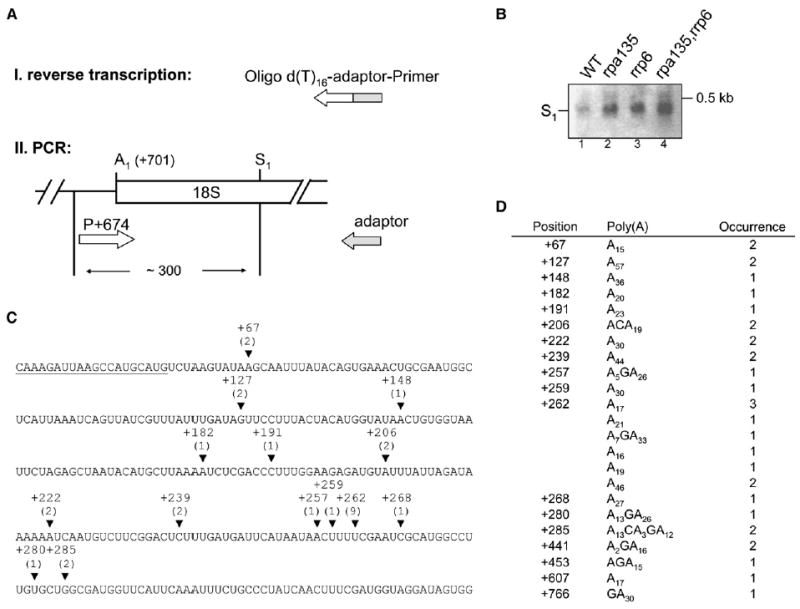
Total RNA from the four strains analyzed in Figure 4 was reverse transcribed with an oligo(dT)-adaptor primer. cDNA was then PCR amplified with the adaptor primer in combination with the forward primer (position +674 relative to the 5′ end of 35S pre-rRNA). PCR products were cloned and sequenced.
(A) Schematic diagram of the procedure and the approximate size of PCR products are indicated.
(B) EtBr-stained gel obtained after agarose gel electrophoresis of PCR products. S1 indicates PCR products derived from RNA formed by cleavage/polyadenylation at S1.
(C) Sequence of 18S rRNA is shown from position +37 to +337 with respect to the 5′ end of mature 18S rRNA. Position of forward primer for amplification of sequencing product is underlined. Polyadenylation sites identified between +441 and +766 are not shown (see table in [D]). Arrowheads indicate the position of polyadenylation sites (from strain NOY2176). If adenosines in the 18S rRNA sequence precede a poly(A) stretch the last nucleotide is indicated. The number of sequenced transformants are given in parentheses.
(D) The table shows a list of transformants analyzed, position of polyadenylation sites, length and composition of poly(A) tails, and number of sequenced transformants. We note that the sequences of poly(A) tails occasionally contained bases other than A. It is not clear whether this use of non-A in poly(A) tails was actually carried out by TRAMP in vivo. Similar observations were recently made for polyadenylation of human rRNA (Slomovic et al., 2006).
