TABLE 2.
Table of parameters
| Parameter | Description | Value | Source |
|---|---|---|---|
| kon, khyd | Dimer on rate constant; MinE-induced MinD-dimer hydrolysis rate | — | Assumed to be fast relative to other processes. |
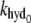 |
Background MinD dimer hydrolysis rate | khyd/10 | Hu et al. (11). |
| kdep | E-ring-induced MinD hydrolysis rate | 80 s−1 | Estimated from Hale et al. (13). |
| kpoly | MinD polymerization rate constant | 100 μM−1 s−1 | Estimated from Hale et al. (13). |
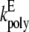 |
E-ring elongation rate constant | 160 μM−1 s−1 | Estimated from Hale et al. (13). |
| knuc | MinD polymer nucleation rate constant | 0.001–0.1 μM−3 s−1 | Tested range; see text. |
| kcap | E-ring initiation rate constant | 0.06–1.5 μM−3 s−1 | Tested range; see text. |
| KE | MinE dimer dissociation constant | 0.6 μM | Zhang et al. (36). |
| Dtot | Total MinD concentration | 4 μM | Estimated from Shih et al. (24) and de Boer et al. (35). |
| Etot | Total MinE concentration | 3 μM | Estimated from Shih et al. (24). |
| Dthresh | Threshold MinD concentration | 1.5–3 μM | Hu et al. (11). |
| Ethresh | Threshold MinE concentration | 2.5 μM | Fitted parameter; see text. |
| δ | Half MinD monomer length | 2.5 nm | Suefuji et al. (25). |
| L, r, V | Cell length, radius, volume | 3 μm, 3/8 μm, 1.3 μm3 | Approximate dimensions. |
| Ap, Am | Polar and nonpolar surface areas | 1 μm2, 10 μm2 | Approximate dimensions. |
| θ | MinD helix pitch angle | 80° | Estimated from Shih et al. (9). |
| λ | Phage expression level | 3.3 μM | Fitted parameter; see text. |
Parameters used in the deterministic and stochastic versions of the model. A more detailed discussion of parameter values is provided in the Appendix.
