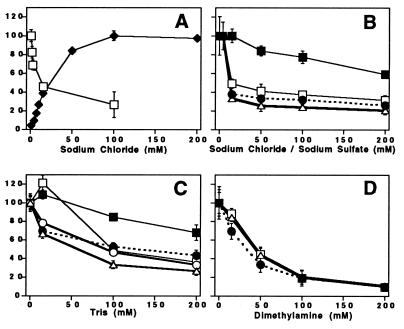
Figure 3.
Effect of various compounds on O2 evolution and 14C labeling of subunits in PSII at pH 7.5. The incubation time with the 14C-labeled amine was 30 min. In some cases, the error bars are smaller than the symbols used to represent the data. In A, solid diamonds, the O2 evolution activity of PSII-2 at pH 7.5 was measured as a function of added NaCl. Sample buffer contained: 0.4 M sucrose/50 mM Hepes-NaOH, pH 7.5/10 mM Ca(OH)2/1 mM potassium ferricyanide/0.25 mM recrystallized 2,6-dichlorobenzoquinone. The maximum (100%) corresponds to 350 μmol O2 (mg Chl-hr)−1. In A, open squares, the amount of 14C bound to CP47 in PSII-2 was measured as a function of added NaCl. Samples were incubated with 0.19 mM [14C]benzylamine-hydrochloride (free base concentration, 2.8 μM). In B, the amount of bound 14C was measured as a function of added NaCl. PSII-2 was incubated with 4 mM [14C]methylamine-hydrochloride (free base concentration, 2.8 μM). Because [14C]methylamine was only available as the hydrochloride, the first data point was obtained at 4 mM Cl−. Data shows labeling of CP47 (open squares), MSP (open triangles), and the D2/D1 polypeptides (solid circles). In B, the amount of bound 14C in CP47 was also measured as a function of added sodium sulfate after incubation of PSII-2 with 4 mM [14C]methylamine-hydrochloride (solid squares). MSP and the D2/D1 polypeptides behaved similarly on the addition of sodium sulfate (data not shown). In C and D, the amount of bound 14C from 4 mM [14C]methylamine-hydrochloride was measured as a function of added (unlabeled) Tris and dimethylamine, respectively. Data shows labeling of CP47 (open squares), MSP (open triangles), and the D2/D1 polypeptides (solid circles) in PSII-2. Data in C also shows the Tris competition with labeling of CP47 (solid squares) and the D2/D1 (open circles) polypeptides in PSII-3.