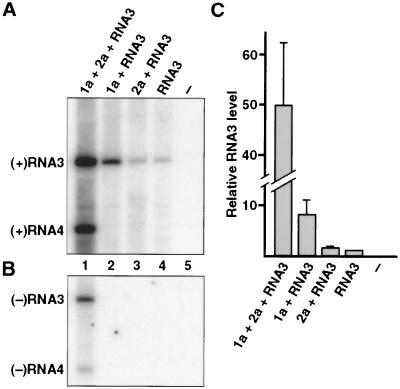
Figure 2.
Northern blot analysis of BMV RNA3 accumulation in yeast expressing the indicated BMV components. To ensure full induction and equilibration of RNA3 transcription from pB3, yeast cells were passaged three times in gal-containing medium and total RNA was extracted. Equal amounts of total RNA per sample were denatured with glyoxal, run on a 0.8% agarose gel, transferred to nylon membranes, and hybridized to 32P-labeled in vitro transcript probes. (A) RNA3-derived positive-strand RNAs detected with a probe complementary to the 3′ 200 bases of BMV RNA3. (B) RNA3-derived negative-strand RNAs, as detected with a probe corresponding to 500 bases of coat protein coding sequence. (C) Positive-strand RNA3 levels in the indicated yeast strains were measured with a Molecular Dynamics PhosphorImager and normalized to the level in yeast expressing RNA3 alone. Averages and standard deviations from five independent experiments are shown.