Figure 2.
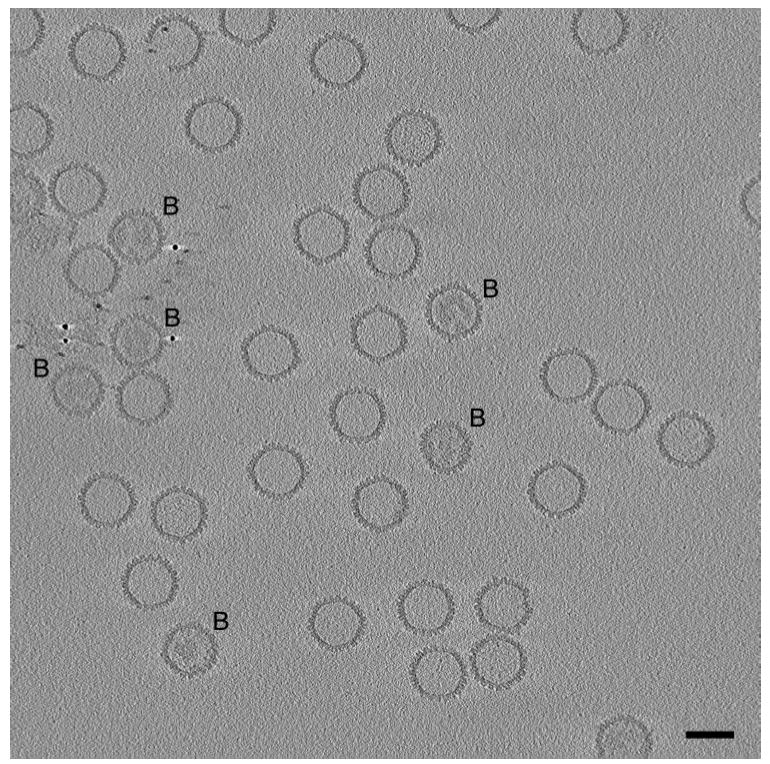
Central slice through a cryo-electron tomogram of a field of HSV-1 capsids. The slice is 3 nm thick. No denoising was applied to the tomogram. Bar=100 nm. Most of the capsids present are A-capsids, i.e. empty capsids. A few are B-capsids (marked B), with more-or-less ordered internal material, representing the scaffolding protein, UL26.5. Several of the gold particles used as fiducial markers are seen in the left/middle of the field. One of the A-capsids (top left) is missing part of its shell. The fact that this slice samples all of the capsids centrally or almost so indicates that they are aligned quite precisely in the z-dimension. Presumably, this alignment results from their being confined within an ice film whose thickness, ~ 150 nm, as measured from the tomogram, is not much greater than the capsid diameter of 125 nm. Most of the capsids are oriented so that they are viewed along or close to a threefold axis of icosahedral symmetry; accordingly, they appear hexagonal in outline, and their icosahedral facets are sampled in slices through the tomogram (not shown) parallel to this one and offset by ~ 50 nm.
