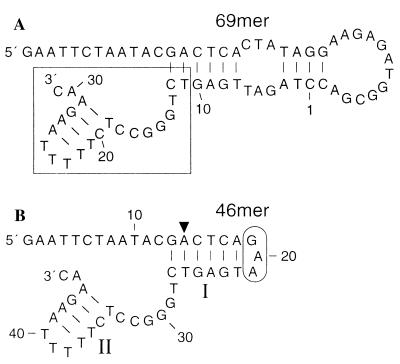
Figure 1.
Sequence and predicted secondary structures of minimized self-cleaving DNAs. (A) Sequence and secondary structure of a synthetic 69-nucleotide self-cleaving DNA that was isolated by in vitro selection. Numbers identify nucleotides that correspond to the 50-nucleotide random-sequence domain that was included in the original DNA pool (note that 19 bases of this domain have been deleted). The conserved nucleotides (nucleotides 11–31, boxed) are similar to those previously used to define this class of deoxyribozymes (16). (B) A 46-nucleotide truncated version of class II DNAs that retains full activity. I and II designate stem–loop structures of the 46-mer that are predicted by the structural folding program dna mfold (18, 19) and that were confirmed by subsequent mutational analysis (Fig. 2). The conserved core of the deoxyribozyme spans nucleotides 27–46 and the major site of DNA cleavage is designated by the arrowhead. Encircled nucleotides can be removed to create a bimolecular complex where nucleotides 1–18 constitute the substrate subdomain, and nucleotides 22–46 constitute the catalyst subdomain.