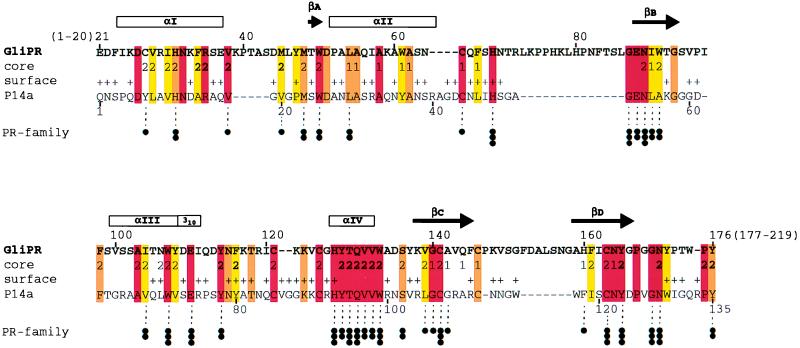
Figure 1.
Sequence alignment of the proteins GliPR and P14a. The one-letter amino acid code is used, and the numbering for GliPR is indicated above its sequence. The locations of the regular secondary structure elements identified for P14a (7) are depicted at the top, with the four α-helices and the 310-helix represented by boxes marked αI, αII, αIII, αIV, and 310, and the four β-strands, by the arrows βA, βB, βC, and βD. In the row “core” the residues forming a small hydrophobic cluster associated with the β-sheet and helix II (Fig. 2 a and b) are indicated 1, those constituting the larger core associated with the β-sheet and helices I, III, and IV (Fig. 2 a and b) with 2, and residues of helix IV and those directly contacting helix IV in P14a (Fig. 2 a and b) with 2. In the row “surface,” + identifies residues that exhibit a solvent accessibility larger than 25% in at least one of the 20 energy-minimized diana conformers of P14a. Color code: residues conserved between GliPR and P14a are orange, and those conserved also within the whole group of known plant PR-1 proteins (see figure 10 in ref. 7) are red. Conserved hydrophobicity of Ala, Val, Leu, Ile, Tyr, Phe, uncharged His, Trp, Pro, Met, or Cys in GliPR and all PR-1 proteins is highlighted in yellow. Conservation within the “PR-protein superfamily” (see text and Fig. 3), derived from multiple alignment of all 26 representative members, is indicated at the bottom: three dots label residues that are strictly conserved within the superfamily, two dots denote conservation of side chain character (hydrophobic: Ala, Val, Leu, Ile, Tyr, Phe, Trp, Met, Cys, and Pro; hydroxyl-bearing: Ser and Thr; or hydrogen bond acceptors: uncharged His, Asp, Asn, Glu, and Gln), and one dot indicates mutations from hydrophobic amino acids to Gly, Thr, or His.