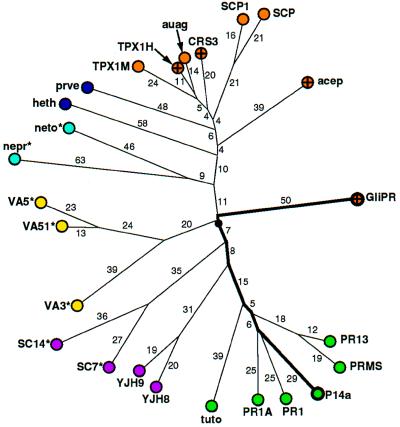
Figure 3.
Genealogical tree of the “PR-protein superfamily,” where the distances are given in PAM units. For clarity, distances with PAM = 1 or 2 are not indicated. Color code: mammals, orange (human proteins are further marked with a cross); plants, green; fungi, magenta; insects, yellow; nematodes, light blue; and other vertebrates, dark blue. The branches are labeled either with the first four letters of the Swiss-Prot database code in capital letters, or with an abbreviation derived from the GenBank and PIR database codes. Proteins for which the putative active site (Fig. 2 c and d and text) is not strictly conserved are identified as follows: where Glu-88 of GliPR is exchanged with Gln there is an asterisk. For “heth” the residue corresponding to His-69 of GliPR is exchanged with Glu. Abbreviations: from GenBank and PIR, acep, acidic epididymal glycoprotein, S80310; auag, autoantigen 1 of the guinea pig sperm acrosome, U35712; heth, helothermine precursor (Mexican beaded lizard), U13619; nemt, gene sequence from chromosome III of nematode Caenorhabditis elegans, Z68108; nepr, nematode protective antigen, A28112; prvn, Prepro-Cys-rich venom protein, U59447; tuto, tumor-related protein from tobacco; from Swiss-Prot, CRS3_HUMAN, PR13_HORVU, PR1A_TOBAC, PR1_ARATH, PRMS_MAIZE, SC14_SCHCO, SC7_SCHCO, SCP1_MOUSE, SCP_RAT, SGP28_X94323, TPX1_HUMAN, TPX1_MOUSE, VA3_SOLIN, VA51_VESCR, VA5_VESPE, VJH8_YEAST, and YJH9_YEAST.