FIG 1.
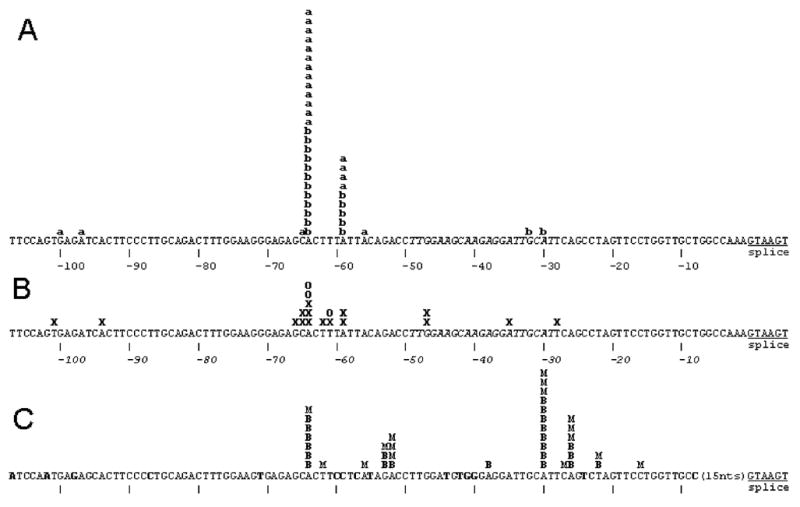
Summary of 5′ termini of NAT2 cDNAs from 5′-RLM-RACE analysis of liver RNA samples and from public databases with comparison to NAT1. A) 5′ termini determined by 5′-RLM-RACE of liver samples from Ambion and BD respectively indicated by “a” and “b”. The positions of the most frequent TSS locations at -59 and -64 are located 8711 and 8716 nucleotides upstream of the NAT2 ORF. B) The 5′ termini of NCBI database clones from liver (D90042, BG569293, AV658623, AV658656, BG533459, BG563731, BG568400, BG618195, BG569272, BC015878, CB161982, CB161860, D90040); colon adenocarcinoma (BX095770, AI792606) and hepatocellular carcinoma (AV684197) are shown as X and the 5′ termini of the small intestine oligo-capped clones HSI05750, HSI08034 and HSI15615 from DBTSS are shown as O. C) NAT1 mRNA termini previously determined by 5′-RLM-RACE with RNA samples from breast and the MCF-7 cell line (Husain et al., 2004) are indicated by B and M respectively. The sequence corresponding to the intron-spanning forward q-RT-PCR primer is shown in italics. In the NAT1 DNA sequence, nucleotides not identical to the NAT2 sequence are shown in bold.
