Abstract
Three hemotropic mycoplasmas have been identified in pet cats: Mycoplasma haemofelis, “Candidatus Mycoplasma haemominutum,” and “Candidatus Mycoplasma turicensis.” The way in which these agents are transmitted is largely unknown. Thus, this study aimed to investigate fleas, ticks, and rodents as well as saliva and feces from infected cats for the presence of hemotropic mycoplasmas, to gain insight into potential transmission routes for these agents. DNA was extracted from arthropods and from rodent blood or tissue samples from Switzerland and from salivary and fecal swabs from two experimentally infected and six naturally infected cats. All samples were analyzed with real-time PCR, and some positive samples were confirmed by sequencing. Feline hemotropic mycoplasmas were detected in cat fleas and in a few Ixodes sp. and Rhipicephalus sp. ticks collected from animals but not in ticks collected from vegetation or from rodent samples, although the latter were frequently Mycoplasma coccoides PCR positive. When shedding patterns of feline hemotropic mycoplasmas were investigated, “Ca. Mycoplasma turicensis” DNA was detected in saliva and feces at the early but not at the late phase of infection. M. haemofelis and “Ca. Mycoplasma haemominutum” DNA was not amplified from saliva and feces of naturally infected cats, despite high hemotropic mycoplasma blood loads. Our results suggest that besides an ostensibly indirect transmission by fleas, direct transmission through saliva and feces at the early phase of infection could play a role in the epizootiology of feline hemotropic mycoplasmas. Neither the investigated tick nor the rodent population seems to represent a major reservoir for feline hemotropic mycoplasmas in Switzerland.
The agent formerly known as Haemobartonella felis has recently been reclassified as a hemotropic mycoplasma, and three different species have been characterized in cats: Mycoplasma haemofelis, “Candidatus Mycoplasma haemominutum,” and “Candidatus Mycoplasma turicensis” (2, 9, 18, 19, 27, 34). Infections with feline hemotropic mycoplasmas can induce a fulminant, potentially fatal hemolytic crisis, but the pathogenic potential varies greatly among the three different species.
Some years ago, sensitive PCR assays became available for the specific diagnosis of feline hemotropic mycoplasmas (2, 6, 13), and real-time PCR assays have been developed which allow the differentiation and quantification of the three species (28, 33, 34). In applying PCR-based methods, feline hemotropic mycoplasma infections in pet cats have been diagnosed worldwide (6, 13, 16, 25, 26, 30, 33, 36), and a recent study has documented infections in 12 different wild felid species from three different continents (35). Nevertheless, the epizootiology of hemotropic mycoplasmas is still poorly understood, and the transmission routes are largely unknown. Experimental transmission via intravenous, intraperitoneal, and oral routes using infected blood has been successful (8). However, several studies indicate that blood-sucking arthropods could represent the natural means of transmission among cats. In dogs, Mycoplasma haemocanis (formerly Haemobartonella canis), a canine hemotropic mycoplasma that is very closely related to M. haemofelis, can successfully be transmitted among dogs via the dog tick Rhipicephalus sanguineus (21). Furthermore, “Ca. Mycoplasma haemominutum” DNA was recently reported in unfed Ixodes ovatus ticks collected from three different areas in Japan (24). Other PCR-based studies demonstrated “Ca. Mycoplasma haemominutum” and M. haemofelis DNA in cat fleas (Ctenocephalides felis) collected from experimentally or naturally infected cats (15, 22, 37), and DNA of both hemotropic mycoplasmas was detected in cat flea feces (37). However, an attempt to experimentally transmit M. haemofelis and “Ca. Mycoplasma haemominutum” between cats via the hematophagous activity of C. felis was not conclusive: only one out of six cats fed on by M. haemofelis-PCR-positive fleas transiently turned PCR positive, and clinical or hematological signs consistent with feline infectious anemia did not develop in the cat (37). Furthermore, none of the cats fed on by “Ca. Mycoplasma haemominutum”-PCR-positive fleas yielded PCR-positive results in the blood, and the attempt to experimentally transmit M. haemofelis or “Ca. Mycoplasma haemominutum” by feeding cats with infected C. felis was not successful (38).
The discovery that “Ca. Mycoplasma turicensis” is most closely related to rodent hemotropic mycoplasmas, namely, Mycoplasma coccoides and Mycoplasma haemomuris, brought up the hypothesis of an interspecies transmission of hemotropic mycoplasmas between rodents and cats (34). In addition, there is evidence for a direct transmission of hemotropic mycoplasmas between cats. In a recent study, “Ca. Mycoplasma haemominutum” but not M. haemofelis was detected by PCR in the saliva and salivary glands of cats experimentally infected with the respective hemotropic mycoplasma (7). Furthermore, male cats and cats with outdoor access were more frequently infected with hemotropic mycoplasmas (17, 25, 33, 36), and a history of cat bite abscesses increased the relative risk for infection (11). Hemotropic mycoplasma infections were even reported in areas where flea or tick infestations are uncommon (13).
The aims of the present study were to investigate fleas, ticks, and rodents as well as saliva and feces from infected cats for the presence of hemotropic mycoplasmas to gain insight into potential transmission routes of these agents.
(These studies were conducted by B. Willi in partial fulfillment of the requirements for a Ph.D. degree at the Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.)
MATERIALS AND METHODS
Arthropods.
A total of 2,198 ticks and 77 fleas were included in the study (Table 1). The 181 ticks from 39 cats and 66 dogs and the 77 fleas from 21 cats were collected by pet owners and veterinarians in northern Switzerland. Because Rhipicephalus sanguineus has been reported as a vector of M. haemocanis (21), a collection of 67 Rhipicephalus sp. ticks was included in the study; these ticks were derived from southern Switzerland because Rhipicephalus sp. is not permanently established north of the Alps. Nucleic acid (NA) from 41 of the latter ticks was extracted during a previous study (3). Additionally, NA was available from 1,950 unfed ticks that had been collected from vegetation in the area around Zurich, Switzerland, by the cloth-dragging method (1), during an unrelated study. The arthropods were stored at −20°C in liquid nitrogen or in ethanol at 4°C until transported to the Clinical Laboratory, University of Zurich, Switzerland. Before NA extraction, the ticks and fleas collected from cats and dogs in northern Switzerland were microscopically identified based on their morphology (5, 31).
TABLE 1.
Arthropod and rodent species investigated for the presence of hemotropic mycoplasmasa
| Sample group | No. of samples | Source(s) or reference | Pooled for extraction | NA extraction method | No. (%) tested PCR positive for the indicated speciese
|
|||
|---|---|---|---|---|---|---|---|---|
| M. haemofelis | “Ca. Mycoplasma haemominutum” | “Ca. Mycoplasma turicensis” | M. coccoides | |||||
| Ticks from animals | ||||||||
| Ixodes sp. | 71 | 39 cats | 1-3 | DNeasy tissue kit | 0 | 2 (2.8) | 0 | ND |
| 110 | 66 dogs | 1-4 | DNeasy tissue kit | 0 | 0 | 0 | ND | |
| Rhipicephalus sp. | ||||||||
| Rhipicephalus sp. | 26 | Miscellaneous | DNeasy tissue kit | 0 | 0 | 0 | ND | |
| R. sanguineus | 23 | 3 | QIAamp DNA minikit | 0 | 0 | 1 (4.3)d | ND | |
| R. turanicus | 18 | 3 | QIAamp DNA minikit | 0 | 0 | 0 | ND | |
| Ticks from vegetation | 1,950 | Vegetation | 10 | MagNa Pure | 0 | 0 | 0 | ND |
| Fleas | ||||||||
| C. felis | 73 | 17 cats | 1-5 | DNeasy tissue kit | 0 | 2 (2.7) | 0 | ND |
| C. canis | 4 | 4 cats | DNeasy tissue kit | 0 | 0 | 0 | ND | |
| Rodents | ||||||||
| A. terrestris | 186 | Free-living rodentsb,c | MagNa Pure | 0 | 0 | 0 | 0 | |
| Apodemus sp. | 45 | Free-living rodents | MagNa Pure | 0 | 0 | 0 | 24 (53) | |
| M. glareolus | 11 | Free-living rodentsc | MagNa Pure | 0 | 0 | 0 | 1 (9.1) | |
| Microtus sp. | 7 | Free-living rodentsc | MagNa Pure | 0 | 0 | 0 | 0 | |
| M. domesticus | 7 | Free-living rodents | MagNa Pure | 0 | 0 | 0 | 0 | |
The table shows the species, number, source, and extraction methods used for ticks, fleas, and rodents collected from throughout Switzerland and the numbers and percentages that tested PCR positive for M. haemofelis, “Ca. Mycoplasma haemominutum,” “Ca. Mycoplasma turicensis,” and M. coccoides.
Reference 20 (Schwarzenbach et al., 2004).
Reference 23 (Stieger et al., 2002).
Collected indoors from a dog-keeping household or directly from a dog.
ND, not determined.
Rodents.
Samples from 256 free-living Swiss rodents were available. Serosanguinous fluid was derived from 187 rodents (184 Arvicola terrestris, 1 Myodes glareolus, and 2 Microtus sp.) from Zurich, Switzerland, which had been caught and specified during other studies (20, 23). In addition, liver and spleen tissue samples were collected from 69 rodents (2 A. terrestris, 45 Apodemus sp., 10 M. glareolus, 5 Microtus sp., and 7 Mus domesticus) from the canton of Grisons, Switzerland; the latter rodents were morphologically specified before necropsy (12).
Salivary and fecal swabs from cats.
From two cats (Cat 1 and Cat 2) experimentally infected with “Ca. Mycoplasma turicensis” (34), salivary and fecal swabs were regularly collected with commercially available cotton swabs (Q-Tips) until days 226 and 186 postinfection (p.i.), respectively. Swabs were inserted into the cheek pouches or the rectums of the cats and then placed into 1.5-ml microcentrifuge tubes. The external tips were removed, and the tubes were closed. In addition, salivary and fecal swabs were collected by veterinarians from privately owned cats infected with “Ca. Mycoplasma haemominutum” (four cats), M. haemofelis (one cat; only a fecal swab was available) or “Ca. Mycoplasma turicensis” (one cat) within 1 day to 4 weeks after the hemotropic mycoplasma PCR-positive result from blood was obtained; swabs were sent to the laboratory within 1 day after collection. All swabs were stored at −20°C until NA extraction.
NA extraction.
Some tick and flea samples were pooled for NA extraction (Table 1); pools consisted of the arthropods of one species and those collected from one animal. The arthropods were mechanically disrupted with sterile scalpel blades and homogenized in a Mixer Mill MM 300 device (Retsch GmbH, Haan, Germany). NA extraction was performed with a DNeasy tissue kit (QIAGEN, Hombrechtikon, Switzerland) or a MagNA Pure LC total nucleic acid isolation kit (Roche Diagnostics, Rotkreuz, Switzerland) (Table 1). Ixodes sp. ticks from cats and dogs, some of them engorged with blood, were weighed, and 180 μl of ATL buffer (supplied in the kit) per 25 mg of weight was added. NA from serosanguinous fluid and tissues from rodents was extracted with a MagNA Pure LC total nucleic acid isolation kit (Roche) and a MagNA Pure LC DNA isolation kit II (Roche), respectively. NA from salivary and fecal swabs was extracted with a MagNA Pure LC total nucleic acid isolation kit (Roche) as described previously (10). During each NA extraction, negative controls consisting of 100 μl phosphate-buffered saline were concurrently prepared with each batch of 15 samples to monitor for cross-contamination.
PCR assays.
As an internal control for the presence of amplifiable NA, samples extracted from ticks, fleas, and rodents were subjected to a real-time PCR assay for 18S rRNA gene amplification (Applied Biosystems, Rotkreuz, Switzerland). Real-time PCR assays based on the 16S rRNA genes of M. haemofelis, “Ca. Mycoplasma haemominutum,” and “Ca. Mycoplasma turicensis” were performed as described previously (33, 34). Ticks from dogs were analyzed with a real-time PCR assay that amplifies both “Ca. Mycoplasma haemominutum” and “Candidatus Mycoplasma haematoparvum” (14). Due to their high sequence similarity, Mycoplasma haemocanis is also amplified in the M. haemofelis real-time PCR assay (33). NA extracted from serosanguinous fluid from rodents was pooled (8 NA samples/pool) for real-time PCR analyses. For the specific detection of Mycoplasma coccoides in rodent samples, oligonucleotides were designed as follows: forward primer McoccF (5′-GAACGATGAAGGTCATTTTGATTG-3′), reverse primer McoccR (5′-CTGGCACATAGTTWGCTGTCACTTA-3′), and probe Mcocc-MGB (6-FAM-AATTATGATGGTACCTCCTG-MGB). The primer McoccR is identical to the reverse primer used in the three feline hemotropic mycoplasma real-time PCR assays (33). In each PCR run, the amplification buffer contained dUTP for use with uracil-N-glycosylase to prevent the carryover of PCR amplicons, and water was used as a sham control. All sham extraction and pipetting controls tested PCR negative.
Sequencing.
In order to confirm the presence of “Ca. Mycoplasma haemominutum” in real-time PCR-positive ticks, approximately 200 bp of the 16S rRNA gene was amplified using primers that amplify canine and feline hemotropic mycoplasmas (13), and amplification products were subjected to direct sequencing. To confirm M. coccoides PCR-positive results, primers (forward primer Mcocc364F [5′-ACGAAAGTCTGATGGAGCAAT-3′] and reverse primer Mcocc533R [5′-ACGCCCAATAAATCCGAATAA-3′]) were used to amplify a 169-bp product of the 16S rRNA gene of the agent. The reaction mixture contained 12.5 μl of 2× qPCR Mastermix (Eurogentec, Seraing, Belgium), 640 nM of each primer, and 5 μl of template DNA, made up to 25 μl with water. The thermal program comprised 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 1 min. The amplification products were visualized by ethidium bromide staining on a 3% agarose gel before being subjected to direct sequencing. The sequences were aligned using CLUSTAL W (29), and the percent identity was calculated with Jalview 2.07 (4).
Statistics.
For observed sample prevalences, 95% confidence intervals (CI) were calculated (WinEpiscope 2.0, Zaragoza, Spain). The frequency of PCR-positive results was compared among Ixodes sp. ticks collected from pet animals and from vegetation by using Fisher's exact test (expected cell frequencies ≤5). Differences were considered significant with P values of <0.05.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene nucleotide sequences generated from M. coccoides isolates have been submitted to GenBank and given the accession numbers EF175168, EF175169, and EF175170.
RESULTS
Sample characteristics.
All ticks collected from pet animals in northern Switzerland were identified as Ixodes sp. (Table 1); the 77 fleas collected from cats included 73 C. felis and 4 Ctenocephalides canis. The species of the 1,950 ticks collected from the vegetation around Zurich had not been microscopically specified. However, based on a previous study (1) and our own experience (32), we assumed that the unspecified ticks consisted mainly of Ixodes ricinus ticks, which were the main species captured by the cloth-dragging method from grassland in this geographical region. All arthropod and rodent samples tested PCR positive for 18S rRNA genes. In some NA samples extracted from ticks from cats and dogs, the 18S rRNA gene assay revealed unexpectedly high threshold cycle (CT) values (≥30), which could be attributable to inhibition of the PCR. Inhibition was confirmed by testing a 1:10 dilution of the samples; while a CT value of roughly 3.5 higher is expected for an uninhibited PCR (sample dilution of 1:10), the CT values obtained from our samples were 13 to 29 CT lower after dilution. These samples were therefore assayed in the PCRs following by using a 1:10 dilution.
Feline hemotropic mycoplasmas in blood-sucking arthropods.
Arthropods were analyzed by real-time PCR for the presence of hemotropic mycoplasma DNA (Table 1). Three ticks and two fleas collected from animals tested positive with real-time PCR for hemotropic mycoplasmas; all positive samples were extracted from individual arthropods. PCR-positive results for “Ca. Mycoplasma haemominutum” were obtained from 2.7% (95% CI, 0 to 6.4%) of the cat fleas and from 2.8% (95% CI, 0 to 6.6%) of the Ixodes sp. ticks collected from Swiss pet cats; both PCR-positive Ixodes sp. ticks were fully engorged with blood before being subjected to NA extraction. “Ca. Mycoplasma turicensis” was found in 1 (4.3%; 95% CI 0 to 12.6%) R. sanguineus tick collected from southern Switzerland. None of the 1,950 ticks collected from vegetation in the region around Zurich tested positive for hemotropic mycoplasma by PCR. Hemotropic mycoplasmas were more frequently detected in Ixodes sp. ticks picked from pet animals than in unfed ticks collected directly from vegetation in Switzerland (P = 0.0144).
To confirm the two “Ca. Mycoplasma haemominutum” PCR-positive results with Ixodes sp. ticks, 171 bp of the 16S rRNA gene was sequenced and aligned with published “Ca. Mycoplasma haemominutum” (GenBank accession no. DQ157149) and “Ca. Mycoplasma haematoparvum” (GenBank accession no. AY532390) sequences; a higher identity was found with “Ca. Mycoplasma haemominutum” (99%) than with “Ca. Mycoplasma haematoparvum” (97% to 98%).
Hemotropic mycoplasmas in rodent samples.
All NA samples extracted from serosanguinous fluid or tissues from rodents in Switzerland tested negative with real-time PCR for the three feline hemotropic mycoplasmas (Table 1). To test whether hemotropic mycoplasmas are common in free-living rodents in Switzerland and could be amplified from these samples, they were subjected to a real-time PCR assay specific for M. coccoides: 24 Apodemus sp. (53%; 95% CI, 38.4 to 67.6%) samples and 1 M. glareolus (9.1%; 95% CI, 0 to 26.1%) sample tested PCR positive. To again confirm the M. coccoides PCR-positive results, 141 bp amplified from three positive samples was aligned with published M. coccoides (GenBank accession no. AY171918), M. haemomuris (GenBank accession no. U82963), and “Ca. Mycoplasma turicensis” (GenBank accession no. DQ157150) 16S rRNA gene sequences; the highest identity was found with M. coccoides (96 to 98%) and to a lesser degree with M. haemomuris (92%) and with “Ca. Mycoplasma turicensis” (88 to 89%).
Feline hemotropic mycoplasmas in salivary and fecal swabs.
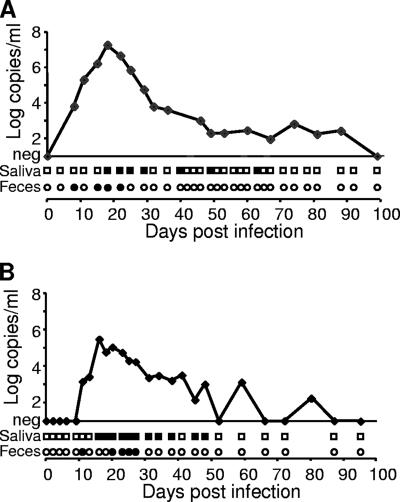
“Ca. Mycoplasma turicensis” DNA was detectable in both salivary and fecal swabs collected from the two experimentally infected cats (Fig. 1). PCR-positive results were obtained from 7 out of 42 salivary swabs collected from Cat 1 (day 18 to 63 p.i.) and from 11 out of 32 salivary swabs collected from Cat 2 (day 16 to 48 p.i.). The CT values ranged from 33 to 37 and from 31 to 39 for Cat 1 and Cat 2, respectively. For the fecal swabs, 4 out of 42 swabs collected from Cat 1 (day 8 to 22 p.i.) and 5 out of 32 swabs collected from Cat 2 (day 11 to 27 p.i.) tested PCR positive. The CT values ranged from 34 to 38 and from 34 to 37 for Cat 1 and Cat 2, respectively. Although saliva and feces were not collected in a quantitative way, a CT value above 31 corresponds to less then 400 copies per PCR. All swabs collected from the two cats at time points with undetectable bacteremia tested PCR negative. Furthermore, all fecal and salivary swabs collected from privately owned pet cats tested negative for hemotropic mycoplasma by PCR, although some cats showed remarkably high hemotropic mycoplasma blood loads (up to 9.7 × 106 copies/ml blood).
FIG. 1.
Kinetics of “Ca. Mycoplasma turicensis” blood loads (curve in x-y diagram) and shedding patterns in saliva (squares beneath x axes) and feces (circles beneath x axes) in two experimentally infected cats, Cat 1 (A) and Cat 2 (B). Blood loads are given in log copy numbers of DNA template per milliliter of blood (adapted from reference 34 with permission). PCR-positive swabs are indicated by filled symbols; negative swabs are depicted by open symbols. Only the first 100 days p.i. are shown; all swabs collected after 100 days p.i. tested PCR negative.
DISCUSSION
This is the first study to report on hemotropic mycoplasma shedding patterns in saliva and feces in infected cats. In addition, it provides a first insight into the occurrence of hemotropic mycoplasmas in arthropods and free-living rodents in Switzerland.
By monitoring two cats experimentally infected with “Ca. Mycoplasma turicensis,” we demonstrated that hemotropic mycoplasma DNA can be detected in saliva and feces up to 9 weeks after infection. Thus, a direct transmission of feline hemotropic mycoplasmas between cats might indeed play a role in the epizootiology of these agents; direct transmission has recently been suggested, based on the common association of hemotropic mycoplasma infection with male gender, outdoor access, and cat bite abscesses (11, 17, 25, 33, 36). “Ca. Mycoplasma turicensis” was not detectable in saliva or feces of experimentally infected cats at later stages of infection. In addition, all feces and saliva samples from privately owned cats tested PCR negative, although some of these cats showed rather high hemotropic mycoplasma blood loads. This finding could indicate that hemotropic mycoplasmas are excreted in the early phase of infection but to a lesser extent by long-term carriers. Since the hemotropic mycoplasma loads in saliva and feces of “Ca. Mycoplasma turicensis”-infected cats were rather low, it may be assumed that oronasal exposure through mutual grooming or sharing of food dishes is hardly sufficient for transmission. Rather, aggressive interactions among cats involving biting might be necessary for a successful direct transmission of hemotropic mycoplasmas. However, experimental transmission studies must be performed to conclusively demonstrate whether direct cat-to-cat transmission plays a role in the epizootiology of feline hemotropic mycoplasmas.
The frequency of hemotropic mycoplasma PCR-positive cat fleas in the present study (2.7%) is lower than the sample prevalence recently reported for cat fleas collected from cats in the United Kingdom (22) and in the United States (15). In the United Kingdom study, 16 to 37% of the fleas tested real-time PCR positive for “Ca. Mycoplasma haemominutum,” whereas the U.S. study reported 3.3% M. haemofelis and 23.9% “Ca. Mycoplasma haemominutum” PCR-positive results. The lower sample prevalence in the present study could be explained by the fact that most fleas were analyzed individually and not in pools, whereas up to 5 or 14 fleas per cat were pooled for extraction in the United Kingdom and the U.S. study, respectively. Furthermore, the cat fleas in the present study were derived from only 17 cats; if the fleas had been pooled per cat before extraction as performed in the studies mentioned, a prevalence of up to 12% (95% CI, 0 to 27.5%) would have resulted. In addition, hemotropic mycoplasma infections are relatively rare in the Swiss pet cat population (33), which would be in agreement with the low number of hemotropic mycoplasma PCR-positive fleas reported in this study.
We only occasionally detected feline hemotropic mycoplasma DNA in ticks from Switzerland, and all of the almost 2,000 unfed ticks collected directly from vegetation tested PCR negative. This suggests that the tick species under investigation play only a marginal role as reservoirs and vectors of feline hemotropic mycoplasmas in Switzerland. A recent study reported the presence of “Ca. Mycoplasma haemominutum” DNA in unfed I. ovatus ticks in Japan (24), suggesting a transstadial transmission of hemotropic mycoplasmas in the latter tick species. I. ricinus and Ixodes hexagonus, but not I. ovatus, have been reported in Switzerland; different Ixodes species may vary in their capability to harbor these agents.
The results obtained so far do not support our hypothesis of an interspecies transmission of hemotropic mycoplasmas between rodents and cats. We had assumed an interspecies transmission because of the close phylogenetic relationship of “Ca. Mycoplasma turicensis” to rodent hemotropic mycoplasmas. Because not all rodent species indigenous to Switzerland were included in this study and because the sample size for some species was rather low, the potential role of rodents in the transmission of feline hemotropic mycoplasmas cannot be definitely ruled out. It should be noted that up to 53% of the samples of the investigated free-living rodent species tested real-time PCR positive for M. coccoides. These results provide the first PCR-based evidence that wild rodents are natural hosts for M. coccoides and that infections with the latter agent are common in at least some rodent species in Switzerland.
In conclusion, neither the tick nor rodent populations investigated seem to play a major role as reservoirs for feline hemotropic mycoplasmas in Switzerland. Remarkably, we detected “Ca. Mycoplasma turicensis” in feces and saliva of infected cats during the early phase of infection. Thus, besides an ostensibly indirect transmission by fleas, future studies should also address the possibility of a direct transmission of feline hemotropic mycoplasmas, ideally by means of experimental transmission studies.
Acknowledgments
We thank P. Deplazes, F. Ehrensperger, and B. Riond for helpful contributions and excellent support. Special thanks go to B. Weibel, T. Meili Prodan, B. Pineroli, and E. Gönczi for excellent laboratory assistance. Laboratory work was performed using the logistics of the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich.
This work was supported by a research grant (Forschungskredit 2002) from the University of Zurich; by the Janggen-Poehn foundation, St. Gallen; by the Roche Research Foundation, Basel; and by Merial GmbH, Germany. R.H.-L. is the recipient of a professorship from the Swiss National Science Foundation (PP00B-102866).
Footnotes
Published ahead of print on 27 April 2007.
REFERENCES
- 1.Aeschlimann, A. 1972. [Ixodes ricinus, Limmeus, 1758 (Ixodoidea: Ixodidae). Preliminary study of the biology of the species in Switzerland.] Acta Trop. 29:321-340. [PubMed] [Google Scholar]
- 2.Berent, L. M., J. B. Messick, and S. K. Cooper. 1998. Detection of Haemobartonella felis in cats with experimentally induced acute and chronic infections, using a polymerase chain reaction assay. Am. J. Vet. Res. 59:1215-1220. [PubMed] [Google Scholar]
- 3.Bernasconi, M. V., S. Casati, O. Peter, and J. C. Piffaretti. 2002. Rhipicephalus ticks infected with Rickettsia and Coxiella in Southern Switzerland (Canton Ticino). Infect. Genet. Evol 2:111-120. [DOI] [PubMed] [Google Scholar]
- 4.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 5.Cotty, A. 1985. Clé de determination des Ixodidae et Amblyommidae de Suisse. Institut de Zoologie, Université de Neuchâtel, Neuchâtel, Switzerland.
- 6.Criado-Fornelio, A., A. Martinez-Marcos, A. Buling-Sarana, and J. C. Barba-Carretero. 2003. Presence of Mycoplasma haemofelis, Mycoplasma haemominutum and piroplasmids in cats from southern Europe: a molecular study. Vet. Microbiol. 93:307-317. [DOI] [PubMed] [Google Scholar]
- 7.Dean, R., C. R. Helps, T. J. Gruffydd-Jones, and S. Tasker. 2005. Use of real-time PCR to detect M. haemofelis and ‘Candidatus Mycoplasma haemominutum’ in the saliva and salivary glands of haemoplasma-infected cats, p. 554. In BSAVA 2005 Proceedings. British Small Animal Veterinary Association, Gloucester, United Kingdom.
- 8.Flint, J. C., M. H. Roepke, and R. Jensen. 1958. Feline infectious anemia. I. Clinical aspects. Am. J. Vet. Res. 19:164-168. [PubMed] [Google Scholar]
- 9.Foley, J. E., and N. C. Pedersen. 2001. ‘Candidatus Mycoplasma haemominutum’, a low-virulence epierythrocytic parasite of cats. Int. J. Syst. Evol. Microbiol. 51:815-817. [DOI] [PubMed] [Google Scholar]
- 10.Gomes-Keller, M. A., E. Gonczi, R. Tandon, F. Riondato, R. Hofmann-Lehmann, M. L. Meli, and H. Lutz. 2006. Detection of feline leukemia virus RNA in saliva from naturally infected cats and correlation of PCR results with those of current diagnostic methods. J. Clin. Microbiol. 44:916-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grindem, C. B., W. T. Corbett, and M. T. Tomkins. 1990. Risk factors for Haemobartonella felis infection in cats. J. Am. Vet. Med. Assoc. 196:96-99. [PubMed] [Google Scholar]
- 12.Hausser, J. 1995. Säugetiere der Schweiz, vol. 103. Springer, Berlin, Germany.
- 13.Jensen, W. A., M. R. Lappin, S. Kamkar, and W. J. Reagan. 2001. Use of a polymerase chain reaction assay to detect and differentiate two strains of Haemobartonella felis in naturally infected cats. Am. J. Vet. Res. 62:604-608. [DOI] [PubMed] [Google Scholar]
- 14.Kenny, M. J., S. E. Shaw, F. Beugnet, and S. Tasker. 2004. Demonstration of two distinct hemotropic mycoplasmas in French dogs. J. Clin. Microbiol. 42:5397-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lappin, M. R., B. Griffin, J. Brunt, A. Riley, D. Burney, J. Hawley, M. M. Brewer, and W. A. Jensen. 2006. Prevalence of Bartonella species, Haemoplasma species, Ehrlichia species, Anaplasma phagocytophilum, and Neorickettsia risticii DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 8:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobetti, R. G., and S. Tasker. 2004. Diagnosis of feline haemoplasma infection using a real-time PCR assay. J. S. Afr. Vet. Assoc. 75:94-99. [DOI] [PubMed] [Google Scholar]
- 17.Luria, B. J., J. K. Levy, M. R. Lappin, E. B. Breitschwerdt, A. M. Legendre, J. A. Hernandez, S. P. Gorman, and I. T. Lee. 2004. Prevalence of infectious diseases in feral cats in northern Florida. J. Feline Med. Surg. 6:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neimark, H., K. E. Johansson, Y. Rikihisa, and J. G. Tully. 2001. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of ‘Candidatus Mycoplasma haemofelis’, ‘Candidatus Mycoplasma haemomuris’, ‘Candidatus Mycoplasma haemosuis’ and ‘Candidatus Mycoplasma wenyonii’. Int. J. Syst. Evol. Microbiol. 51:891-899. [DOI] [PubMed] [Google Scholar]
- 19.Rikihisa, Y., M. Kawahara, B. Wen, G. Kociba, P. Fuerst, F. Kawamori, C. Suto, S. Shibata, and M. Futohashi. 1997. Western immunoblot analysis of Haemobartonella muris and comparison of 16S rRNA gene sequences of H. muris, H. felis, and Eperythrozoon suis. J. Clin. Microbiol. 35:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzenbach, G. A., D. Hegglin, C. Stieger, P. Deplazes, and P. I. Ward. 2004. An experimental field approach to parasitism and immune defence in voles. Parasitology 129:93-99. [DOI] [PubMed] [Google Scholar]
- 21.Seneviratna, P., Weerasinghe, and S. Ariyadasa. 1973. Transmission of Haemobartonella canis by the dog tick, Rhipicephalus sanguineus. Res. Vet. Sci. 14:112-114. [PubMed] [Google Scholar]
- 22.Shaw, S. E., M. J. Kenny, S. Tasker, and R. J. Birtles. 2004. Pathogen carriage by the cat flea Ctenocephalides felis (Bouche) in the United Kingdom. Vet. Microbiol. 102:183-188. [DOI] [PubMed] [Google Scholar]
- 23.Stieger, C., D. Hegglin, G. Schwarzenbach, A. Mathis, and P. Deplazes. 2002. Spatial and temporal aspects of urban transmission of Echinococcus multilocularis. Parasitology 124:631-640. [DOI] [PubMed] [Google Scholar]
- 24.Taroura, S., Y. Shimada, Y. Sakata, T. Miyama, H. Hiraoka, M. Watanabe, K. Itamoto, M. Okuda, and H. Inokuma. 2005. Detection of DNA of ‘Candidatus Mycoplasma haemominutum’ and Spiroplasma sp. in unfed ticks collected from vegetation in Japan. J. Vet. Med. Sci. 67:1277-1279. [DOI] [PubMed] [Google Scholar]
- 25.Tasker, S., S. H. Binns, M. J. Day, T. J. Gruffydd-Jones, D. A. Harbour, C. R. Helps, W. A. Jensen, C. S. Olver, and M. R. Lappin. 2003. Use of a PCR assay to assess the prevalence and risk factors for Mycoplasma haemofelis and ‘Candidatus Mycoplasma haemominutum’ in cats in the United Kingdom. Vet. Rec. 152:193-198. [DOI] [PubMed] [Google Scholar]
- 26.Tasker, S., J. A. Braddock, R. Baral, C. R. Helps, M. J. Day, T. J. Gruffydd-Jones, and R. Malik. 2004. Diagnosis of feline haemoplasma infection in Australian cats using a real-time PCR assay. J. Feline Med. Surg. 6:345-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tasker, S., C. R. Helps, C. J. Belford, R. J. Birtles, M. J. Day, A. H. Sparkes, T. J. Gruffydd-Jones, and D. A. Harbour. 2001. 16S rDNA comparison demonstrates near identity between an United Kingdom Haemobartonella felis strain and the American California strain. Vet. Microbiol. 81:73-78. [DOI] [PubMed] [Google Scholar]
- 28.Tasker, S., C. R. Helps, M. J. Day, T. J. Gruffydd-Jones, and D. A. Harbour. 2003. Use of real-time PCR to detect and quantify Mycoplasma haemofelis and “Candidatus Mycoplasma haemominutum” DNA. J. Clin. Microbiol. 41:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe, M., M. Hisasue, K. Hashizaki, M. Furuichi, M. Ogata, S. Hisamatsu, E. Ogi, M. Hasegawa, R. Tsuchiya, and T. Yamada. 2003. Molecular detection and characterization of Haemobartonella felis in domestic cats in Japan employing sequence-specific polymerase chain reaction (SS-PCR). J. Vet. Med. Sci. 65:1111-1114. [DOI] [PubMed] [Google Scholar]
- 31.Weidner, H. 2005. Bestimmungstabellen der Vorratsschädlinge und des Hausungeziefers Mitteleuropas. Anz. Schädlingskd. 44:61. [Google Scholar]
- 32.Wicki, R., P. Sauter, C. Mettler, A. Natsch, T. Enzler, N. Pusterla, P. Kuhnert, G. Egli, M. Bernasconi, R. Lienhard, H. Lutz, and C. M. Leutenegger. 2000. Swiss Army Survey in Switzerland to determine the prevalence of Francisella tularensis, members of the Ehrlichia phagocytophila genogroup, Borrelia burgdorferi sensu lato, and tick-borne encephalitis virus in ticks. Eur. J. Clin. Microbiol. Infect. Dis. 19:427-432. [DOI] [PubMed] [Google Scholar]
- 33.Willi, B., F. S. Boretti, C. Baumgartner, S. Tasker, B. Wenger, V. Cattori, M. L. Meli, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2006. Prevalence, risk factor analysis, and follow-up of infections caused by three feline hemoplasma species in cats in Switzerland. J. Clin. Microbiol. 44:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willi, B., F. S. Boretti, V. Cattori, S. Tasker, M. L. Meli, C. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2005. Identification, molecular characterization, and experimental transmission of a new hemoplasma isolate from a cat with hemolytic anemia in Switzerland. J. Clin. Microbiol. 43:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willi, B., C. Filoni, J. Catão-Dias, V. Cattori, M. Meli, A. Vargas, F. Martines, M. Roelke-Parker, M.-P. Ryser-Degiorgis, H. Lutz, and R. Hofmann-Lehmann. 2007. Worldwide occurrence of feline hemoplasma infections in different wild felid species. J. Clin. Microbiol. 45:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willi, B., S. Tasker, F. S. Boretti, M. G. Doherr, V. Cattori, M. L. Meli, R. G. Lobetti, R. Malik, C. E. Reusch, H. Lutz, and R. Hofmann-Lehmann. 2006. Phylogenetic analysis of “Candidatus Mycoplasma turicensis” isolates from pet cats in the United Kingdom, Australia, and South Africa, with analysis of risk factors for infection. J. Clin. Microbiol. 44:4430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woods, J. E., M. M. Brewer, J. R. Hawley, N. Wisnewski, and M. R. Lappin. 2005. Evaluation of experimental transmission of Candidatus Mycoplasma haemominutum and Mycoplasma haemofelis by Ctenocephalides felis to cats. AJVR 66:1008-1012. [DOI] [PubMed] [Google Scholar]
- 38.Woods, J. E., N. Wisnewski, and M. R. Lappin. 2006. Attempted transmission of Candidatus Mycoplasma haemominutum and Mycoplasma haemofelis by feeding cats infected Ctenocephalides felis. Am. J. Vet. Res. 67:494-497. [DOI] [PubMed] [Google Scholar]