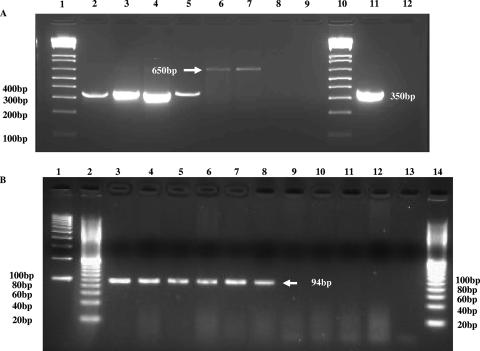
FIG. 3.
PCR results for E. sakazakii zpx using primers BAM 123 and BAM 122 (A) or Es-ProF and Es-ProR (B) for E. sakazakii and non-E. sakazakii strains. Panel A represents a typical 1% agarose gel after electrophoresis, showing the PCR amplification of the 350-bp amplicon. Lanes 1 and 10, Bio-Rad 100-bp molecular size DNA markers; lanes 2 to 5, E. sakazakii strains 11, 569, 573, and 665, respectively; lanes 6 to 9, non-E. sakazakii strains Enterobacter cloacae strain 23355, Citrobacter freundii strain 7, Klebsiella pneumoniae strain 8, and Escherichia coli strain 4, respectively; lane 11, E. sakazakii strain 29544; lane 12, distilled water was used instead of DNA template. Note that the 650-bp product was sequenced and was found not to be related to that of the metalloprotease gene. Panel B represents a typical 3% agarose gel showing the PCR amplification of the 94-bp band. Lanes 1 and 2, Bio-Rad 100-bp and 20-bp DNA molecular size markers, respectively; lanes 3 to 8, E. sakazakii strains 29544, 4963-71, 1121-73, 9363-75, 570, and 564, respectively; lane 9, distilled water; lanes 10 to 14, negative reactions of Enterobacter cloacae subspecies dissolvens ATCC strain 23373, Enterobacter gergoviae ATCC strain 33028, P. carotovorum strain EC15 (provided by Arvind Bhagwat, USDA, Beltsville, MD), Citrobacter freundii ATCC strain 43864, and 20-bp DNA molecular size marker, respectively.