Abstract
Acute enteric infections caused by salmonellas remain a major public health burden worldwide. Poultry, particularly chickens, are known to be the main reservoir for this zoonotic pathogen. Although some progress has been made in reducing Salmonella colonization of broiler chickens by using biosecurity and antimicrobials, it still remains a considerable problem. The use of host-specific bacteriophages as a biocontrol is one possible intervention by which Salmonella colonization could be reduced. A total of 232 Salmonella bacteriophages were isolated from poultry farms, abattoirs, and wastewater in 2004 and 2005. Three phages exhibiting the broadest host ranges against Salmonella enterica serotypes Enteritidis, Hadar, and Typhimurium were characterized further by determining their morphology and lytic activity in vitro. These phages were then administered in antacid suspension to birds experimentally colonized with specific Salmonella host strains. The first phage reduced S. enterica serotype Enteritidis cecal colonization by ≥4.2 log10 CFU within 24 h compared with controls. Administration of the second phage reduced S. enterica serotype Typhimurium by ≥2.19 log10 CFU within 24 h. The third bacteriophage was ineffective at reducing S. enterica serotype Hadar colonization. Bacteriophage resistance occurred at a frequency commensurate with the titer of phage being administered, with larger phage titers resulting in a greater proportion of resistant salmonellas. The selection of appropriate bacteriophages and optimization of both the timing and method of phage delivery are key factors in the successful phage-mediated control of salmonellas in broiler chickens.
Salmonella continues to be a major public health burden worldwide (http://www.who.int/mediacenter/factsheets/fs139/en/). More than 35,000 cases of human salmonellosis were reported in the United States in 2004 alone (6), and more than 192,000 cases were reported in the European Union during the same period (13). The annual cost of medical treatment for salmonellosis, in addition to lost productivity, imposes a considerable financial burden on many countries. The USDA estimated this cost at more than $2.3 billion for the United States in 2005 (http://www.ers.usda.gov/briefing/FoodborneDisease/features.htm). Contaminated poultry products are widely accepted as a major source of Salmonella infections (7). However, controlling Salmonella in poultry is problematic and for broiler chickens this has relied historically on a combination of farm biosecurity and the use of antibiotics (11). Concerns regarding the use of chemical additives in food production have led the European Union to ban many of the antibiotics and growth promoters used in the rearing of broiler chickens. These include spiramycin and tylosin phosphate, which were banned in 1999 (4). In the United States, the FDA has taken similar steps with the recent withdrawal of enrofloxacin for use in poultry production (http://www.fda.gov/oc/antimicrobial/baytril.pdf). Banning or significantly reducing agricultural antibiotic usage may reduce produce quality, yields, and microbiological safety (26). Likewise, constraints on chemical treatments such as chlorine in the abattoir may increase the risk of contamination with bacterial pathogens. Further, significant improvements in biosecurity on poultry farms are likely to be very expensive and difficult to maintain (11), so there is a need to find an acceptable, cost-effective way of preventing infection of poultry with Salmonella (2). Bacteriophage therapy is one possible method of achieving this goal which has gained prominence in recent years (25, 27). Bacteriophages (phages) are natural predators of bacteria and are ubiquitous in the environment (21). The use of host-specific bacteriophages has been promoted as a cost-effective and adaptable approach to control zoonotic bacteria (26). Phages have unique advantages compared with antibiotics (17). They replicate only on the targeted subset of bacteria, avoiding the imbalance of commensal gut flora (dysbiosis) often caused by broad-spectrum antibiotics. Additionally, they only replicate as long as the targeted bacterium is present and so are naturally self-limiting (8). Phages have been used against zoonotic pathogens in live animals and on food surfaces in previous studies (3, 5, 24). Bacteriophages have been used to reduce the numbers of Campylobacter jejuni bacteria in commercial broilers by up to 5.0 log10 CFU g−1 cecal content (18). However, only modest reductions of up to 1.3 log10 CFU have been recorded for similar studies with Salmonella enterica serotype Enteritidis (23). Both Campylobacter and Salmonella phages can be isolated readily from poultry excreta and the poultry farm environment (9, 12, 14) and therefore would not introduce any new biological entity into the food chain if used therapeutically. The emergence of bacteriophage-insensitive mutants (BIMs) has long been perceived as a major limitation of phage therapy (8). However, unlike chemotherapeutic agents such as antibiotics, phages constantly evolve to circumvent their host's defenses and resistant bacteria are often less fit or less virulent than their phage-sensitive counterparts (24). Here we describe the use of Salmonella phages to reduce the numbers of different Salmonella serotypes colonizing the ceca of commercial broiler chickens. We also describe the in vitro experiments used to characterize and select suitable phage therapy candidates. Finally, we report the incidence of phage resistance in vivo and the implications of this for Salmonella phage therapy in broiler chickens.
MATERIALS AND METHODS
Isolation of bacteriophage from environmental samples.
Samples of poultry excreta and effluent were taken from 26 farms, poultry-processing plants, and wastewater treatment plants in southern England during 2004 and 2005. These environmental samples were diluted 1:9 (wt/vol) in nutrient broth (NB, CM0001; Oxoid, Basingstoke, United Kingdom) before the addition of 8.0 log10 CFU of each of three Salmonella Nalr host strains (S. enterica serotypes Enteritidis P125109, Hadar 18, and Typhimurium 4/74) and incubation at 37°C for 24 h. Following enrichment, a 1-ml sample of each culture was subjected to centrifugation at 13,000 × g for 5 min. The supernatant was then filtered through a 0.45-μm-pore-size membrane (16533K; Sartorius, Göttingen, Germany) before 20-μl volumes of filtrate were spotted onto the surfaces of Salmonella lawns prepared by a modification of the method described by Sambrook and colleagues (22). Briefly, this consisted of adding 0.1 ml of an 8.0-log10-CFU-ml −1 suspension of Salmonella in maximum-recovery diluent (MRD, CM0733; Oxoid) to 5 ml of molten overlay agar (NB containing 0.5% [wt/vol] bacteriological agar LP0011 [Oxoid]), gently shaking the mixture, and adding it to prewarmed (37°C, 30 min) nutrient agar (CM0003; Oxoid) plates containing 25 μg ml sodium nalidixate−1 (N4382; Sigma, Dorset, United Kingdom) and 1 μg ml novobiocin−1 (N1268; Sigma). The plates were incubated at 37°C for 24 h before examination for plaques. Individual plaques were extracted from the overlay agar with a pipette and suspended in 100 μl of SM buffer (50 mM Tris-Cl [pH 7.5], 0.1 M NaCl, 8 mM MgSO4 · 7H2O, 0.01% gelatin, reagents from Sigma). This suspension was incubated for 15 min at 37°C with 100 μl of an 8.0-log10 suspension of Salmonella before addition to 5 ml of molten overlay agar and pouring onto a nutrient agar plate. These plates were incubated as described above and examined for plaques after 24 h. Single plaques were propagated in this way a total of three times to ensure that the isolates represented a single clone.
Enumeration of salmonellas in cecal contents.
A suspension of cecal contents (1:9, wt/vol) was prepared in MRD and then decimally diluted in the same medium down to 10−4. A 100-μl volume of each dilution was spread plated onto modified brilliant green (BG) agar, (CM0329; Oxoid) containing 25 μg ml sodium nalidixate−1 and 1 μg ml novobiocin−1. The plates were incubated at 37°C for 24 h before typical Salmonella colonies were counted. Representative Salmonella colonies were confirmed by slide agglutination tests with poly(O)-, poly(H)-, and serotype-specific antisera (Pro-Lab Diagnostics, Cheshire, United Kingdom).
Bacteriophage enumeration in cecal contents.
A suspension of cecal contents (1:9, wt/vol) was prepared in SM buffer and subjected to centrifugation at 13,000 × g for 5 min to remove bulk debris. The supernatant was then filtered through a 0.45-μm-pore-size filter to remove any remaining bacteria. Decimal dilutions of this filtrate were prepared in SM buffer down to 10−4 and subsequently spotted (20 μl) onto lawns of the appropriate Salmonella host as described above.
Host range determination.
The host range of each bacteriophage isolate was determined with 70 Salmonella isolates. The Salmonella serotypes and strains used were Enteritidis PT4 (n = 15), Enteritidis UT (n = 4), Enteritidis PT21B (n = 2), Enteritidis PT1B, 4,12:d:− (n = 7), Enteritidis UT (n = 6), Typhimurium PT36 (n = 2), Typhimurium PT208, Typhimurium F98, Binza (n = 5), 3,15:y:− (n = 3), Virchow (n = 2), Typhi, Stanley, Ohio, Kisarowe, Hadar PT2, Togba, Barielly, Munster/Orion, Infantis, Senftenburg, Montevideo, Kattburg, Saint Paul, Kubacha, Amsterdam, Hadar 18, Java, Derby, Braenderup, Agama, and Amina. Bacterial lysis was determined by spotting 20 μl of a 7.0-log10-PFU-ml−1 suspension of phage onto lawns of Salmonella prepared as described above. After allowing 20 min for the spots to be absorbed, the plates were inverted and incubated for 24 h at 37°C before the degree of lysis was recorded.
Phage replication in vitro.
A 20-ml volume of NB, prewarmed at 37°C, was inoculated with 100 μl of an overnight culture of Salmonella (approximately 109 CFU ml−1). The inoculated broth was then incubated statically for 4 h at 37°C. Following incubation, aliquots (200 μl) of this broth were inoculated with dilutions of phage suspension to give multiplicities of infection (MOIs) of approximately 10°, 103, and 106. Three replicate aliquots of each MOI, along with Salmonella-only controls, were then transferred to the wells of a honeycomb microtiter plate. These plates were incubated at 37°C in a Bioscreen-C automated microbiology growth curve analysis system (Labsystems Corp., Finland). The optical density (600 nm) of each well was recorded at 30-min intervals for 24 h. Duplicate samples of each MOI and control were used for the contemporaneous enumeration of Salmonella bacteria and phages in the suspension at 1-h intervals for 10 h and again at 24 h. For enumeration of salmonellas, decimal dilutions of each suspension were spread plated (100 μl) onto BG agar in triplicate and incubated at 37°C for 24 h before examination for Salmonella colonies. For bacteriophage enumeration, a 1-ml aliquot of each suspension was subjected to centrifugation at 13,000 × g for 5 min. The supernatant was then filtered through a 0.45-μm-pore-size filter to remove any remaining bacteria. Decimal dilutions of this filtrate were prepared in SM buffer and subsequently spotted (20 μl) onto lawns of the appropriate Salmonella host as described above.
Examination of phage morphology (electron microscopy).
Eight microliters of an 8.0-log10-PFU-ml−1 suspension of phage was added to the surface of a glow-discharged, carbon-coated Pioloform grid and fixed for 2 min with glutaraldehyde vapor. Excess sample was removed, and the grid was washed with a drop of double-distilled water. Negative staining was performed by adding 1 drop of 0.5% uranyl acetate to the grid surface, and excess stain was removed immediately. The grids were allowed to air dry for 20 min and were then observed with a JEOL 1220 transmission electron microscope. Digital images from the microscope were captured with a SIS Megaview III camera.
Experimental birds.
Salmonella-free Ross broiler chickens were obtained at 34 days of age from a commercial supplier (Lloyd Maunder, Devon, United Kingdom). The birds were housed in groups of three in floor boxes in a controlled environment under strict conditions of biosecurity. To ensure that the experimental birds remained free of naturally occurring infection, fecal samples were taken each day and tested for Salmonella by enrichment in modified Rappaport-Vassiliadis soya peptone broth (CM0669; Oxoid) and then streaking onto BG agar. Fecal samples were also taken to determine if any preexisting Salmonella phages were present by using the enrichment method described for the environmental samples. Following infection, the birds were sacrificed at intervals and the ceca were aseptically removed. The contents of the lumen were collected in sterile universal tubes for Salmonella and phage enumeration. Salmonella colonization of the livers of the birds was ascertained by inserting a swab into the liver and using this to inoculate a BG agar plate for a semiquantitative count. The swab was then enriched for Salmonella in modified Rappaport-Vassiliadis soya peptone broth, followed by plating onto BG agar as described above.
Bacteriophage therapy trials.
Broiler chickens (n = 216 for trial 1 and 108 for trial 2) were separated at random into one of three units. Each unit was used for one of three Salmonella strains: Enteritidis P125109, Hadar 18, or Typhimurium 4/74. The birds in each unit were separated equally into four rooms (A, B, C, or D) and housed in groups of three in floor boxes. The birds in each room were treated as follows: the birds in room A were all inoculated with Salmonella only; in room B, the birds were inoculated with phage only; and in rooms C and D, the birds received both Salmonella and phage. All Salmonella and bacteriophage suspensions were administered by oral gavage. At 36 days of age, the birds in rooms A, C, and D were challenged with 1 ml of an 8.0-log10-CFU-ml−1 suspension of Nalr Salmonella in phosphate-buffered saline (PBS, BR0014; Oxoid); the birds in room B were inoculated with 1 ml of PBS. At 38 days of age, the birds in groups B, C, and D were inoculated with either 1 ml of 9.0 (trial 1) or 11.0 (trial 2) log10 PFU of bacteriophage φ151 (S. enterica serotype Enteritidis P125109), φ25 (S. enterica serotype Hadar 18), or φ10 (S. enterica serotype Typhimurium 4/74) in PBS containing 30% (wt/vol) CaCO3 as an antacid. At the same time, the birds in group A were inoculated with 1 ml of PBS containing 30% (wt/vol) CaCO3. Three animals from each room were sacrificed daily following phage treatment for up to 6 days (trial 1) or 3 days (trial 2). The cecum was aseptically removed from each bird, and the contents were decimally diluted in MRD or SM buffer for the enumeration of salmonellas and phages, respectively.
Statistical treatment of data.
The significance of differences between control and phage-treated experimental groups was determined on log10-transformed data by a single-factor analysis of variance (ANOVA; Microsoft Excel 2002). The significance of increased phage resistance in Salmonella colonies isolated from phage-treated birds was determined by the chi-square test (SPSS 14.0 for Microsoft Windows).
RESULTS
Isolation and characterization of phages.
A total of 232 Salmonella phages were isolated from 26 sampling sites (broiler farms, poultry abattoirs, and wastewater plants) during 2004 and 2005. Following three rounds of serial plaque purification, the host range of each phage was determined against 70 Salmonella isolates representing 24 different serotypes (a portion of this screening is shown in Table 1). The 232 phage isolates could be ascribed to more than 80 lytic profiles. However, when differences between the lytic spectra of bacteriophage are small, further characterization is required to ensure that the phage isolates are not identical. On the basis of the lytic-spectrum data, three phages with the broadest host range against S. enterica serotypes Enteritidis (φ151), Hadar (φ25), and Typhimurium (φ10) were selected for further characterization. Examination of the phage suspensions by electron microscopy revealed that S. enterica serotype Enteritidis phage φ151 belongs to the Myoviridae family of double-stranded DNA phages. S. enterica serotype Hadar phage φ25 and S. enterica serotype Typhimurium phage φ10 belong to the Siphoviridae family of double-stranded DNA phages. Representative electron micrographs of these phages are presented in Fig. 1.
TABLE 1.
Lytic spectra of 10 Salmonella bacteriophage isolates determined on 41 Salmonella host strainsa
| S. enterica serotype or strain | Lysis by bacteriophage:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| φ10 | φ25 | φ27 | φ28 | φ36 | φ37 | φ51 | φ92 | φ104 | φ151 | |
| 3,15:y:− | − | − | − | − | − | − | − | − | − | − |
| 4,12:d:− | +++ | − | − | − | (++) | − | − | (++) | − | (++) |
| Agama | − | − | − | − | − | − | − | − | − | − |
| Amina | − | − | − | − | − | − | − | − | − | − |
| Amsterdam | +++, H | − | − | − | − | − | − | − | − | − |
| Barielly | − | − | − | − | − | − | − | − | − | − |
| Binza | (+++) | − | − | − | − | − | − | − | − | +++ |
| Braenderup | − | − | − | − | − | − | − | − | − | − |
| Derby | +++, H | (+++) | (+++) | (+++) | − | (+++) | − | S | − | +++ |
| Enteritidis (Harrington) | (+++), H | (+++) | (++) | (+++) | (++) | (+++) | (++) | (++) | (++) | S |
| Enteritidis (Platten) | (+++), H | (+++) | (++) | (+++) | (++) | (+++) | ++, H | (++) | ++, H | +++ |
| Enteritidis P125109 | +++, H | (+++) | (++) | (+++) | ++ | (+++) | ++, H | (+++) | ++, H | +++ |
| Enteritidis PT1B | (+++) | (+++) | ++ | (+++) | +++ | (+++) | ++ | +++ | ++ | +++ |
| Enteritidis PT21B | − | (+++) | ++ | (+++) | ++ | (+++) | − | ++ | − | +++ |
| Enteritidis PT4 | (+++) | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Enteritidis UT | (+++) | (+++) | − | (+++) | ++ | (+++) | − | +++ | − | +++ |
| Hadar 18 | − | +++ | ++ | +++ | S | +++ | − | S | − | S |
| Hadar PT2 | − | +++ | +++ | +++ | +++ | +++ | − | ++ | − | − |
| Infantis | − | − | − | − | − | − | − | − | − | − |
| Java | +++, H | (+++) | (++) | (+++) | S | (+++) | − | S | − | +++ |
| Kattburg | − | +++ | ++ | +++ | +++ | +++ | − | +++ | − | − |
| Kisarowe | − | − | − | − | − | − | − | − | − | − |
| Kubacha | − | − | − | − | − | − | − | − | − | − |
| Montevideo | − | − | − | − | − | − | − | − | − | − |
| Munster/Orion | +++, H | − | − | − | − | − | − | − | − | − |
| Ohio | − | − | − | − | − | − | − | − | − | − |
| Saint Paul | (+++) | ++ | − | (+++) | (+++) | (+++) | − | ++ | − | (+++) |
| Senftenburg | − | ++ | + | ++ | − | ++ | − | ++ | − | − |
| Stanley | +++ | +++ | ++ | +++ | +++ | +++ | − | +++ | − | (+) |
| Togba | − | − | − | − | − | − | − | − | − | − |
| Typhi | (+++) | − | − | − | − | − | − | − | − | − |
| Typhimurium (Mabbott) | +++ | (++) | − | (+++) | − | (++) | − | + | − | ++ |
| Typhimurium (Rawlings) | +++ | (+++) | (+++) | (+++) | ++ | (+++) | ++, H | ++ | ++, H | +++ |
| Typhimurium (Turner) | +++ | (+++) | − | (+++) | − | S | − | + | − | +++ |
| Typhimurium 4/74 | +++ | (+++) | (++) | (+++) | (++) | (+++) | − | (++) | − | +++ |
| Typhimurium DT104 | +++ | − | − | − | − | − | − | − | − | (+++) |
| Typhimurium F98 | +++ | (+++), R | (++) | (+++) | S | (+++) | − | S | − | (+++) |
| Typhimurium LT2 | +++, H | (+++) | (++) | (+++) | − | (+++) | − | − | − | +++ |
| Typhimurium PT208 | +++ | (+++) | (++) | (+++) | (++) | (+++) | − | (++) | − | S |
| Typhimurium PT36 | +++ | (+++) | ++ | S, R | (++) | (+++) | ++ | (++) | ++ | +++ |
| Virchow | − | − | − | − | − | − | − | − | − | − |
| Total no. of strains lysed | 24 | 23 | 19 | 22 | 18 | 22 | 7 | 19 | 7 | 20 |
Shown is a subset of the lytic-spectrum data obtained for 232 bacteriophage isolates screened against 70 Salmonella strains. Results were recorded as follows: +++, confluent lysis; ++, semiconfluent lysis; +, individual plaques; parentheses, opalescent lysis; H, halo; R, regrowth of Salmonella; S, shadow lysis; −, no lysis.
FIG. 1.
Electron photomicrographs of Salmonella phages φ10 (A), φ25 (B), and φ151 (C). Phages φ10 and φ25 both exhibit icosahedral heads and flexible tails typical of members of the Siphoviridae family. With its icosahedral head and contractile tail, phage φ151 typifies members of the Myoviridae family. The bars represent 250 nm.
Replication dynamics of phage in vitro.
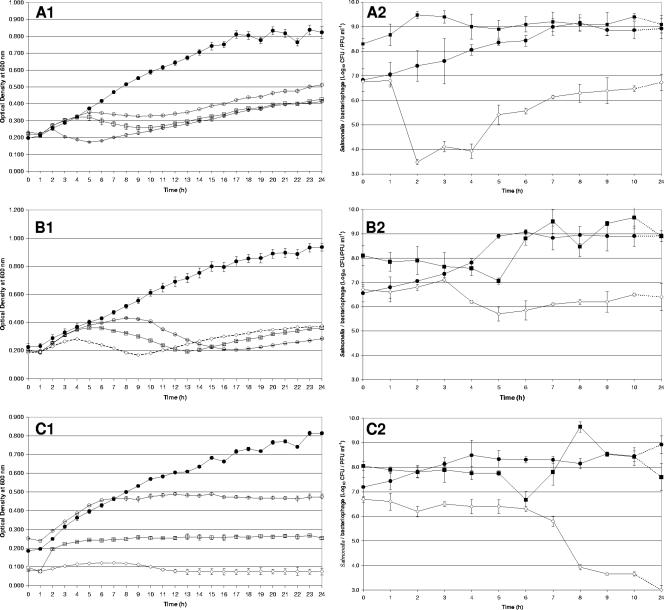
The replication dynamics of each candidate phage were determined in vitro at MOIs of 10°, 103, and 106 prior to application in vivo. Both the optical density and plate count data indicated that the number of bacteria of each Salmonella serotype was reduced appreciably in the presence of phage at each MOI after 24 h (Fig. 2). When the highest MOI was used, S. enterica serotype Enteritidis P12509, Hadar 18, and Typhimurium 4/74 numbers were reduced by means of 2.2, 2.5, and 5.9 log10 CFU ml−1, respectively, after 24 h compared with the controls. These reductions were significant by a single-factor ANOVA (P < 0.00001). The scale and duration of Salmonella reduction varied according to the MOI used, with higher ratios of phage to bacteria resulting in the greatest initial falls (≤10 h following infection). However, the differences between MOI groups generally became less pronounced during the 24-h incubation period (Fig. 2).
FIG. 2.
Graphs A1, B1, and C1 show the optical density readings (at 600 nm) of NB cultures of S. enterica serotypes Enteritidis P125109, Hadar 18, and Typhimurium 4/74, respectively, over a 24-h period. Phages φ151, φ25, and φ10 were separately added to exponential growth phase cultures of S. enterica serotypes Enteritidis, Hadar, and Typhimurium, respectively, at MOIs of 10° (○), 103 (□), and 106 (⋄). Optical density readings taken from uninfected cultures are also shown (•). Graphs A2, B2, and C2 show the plate counts of bacteriophages (▪) and uninfected (•) and infected (⋄) Salmonella cultures recorded from the same samples (only data for the highest MOIs are presented). All means and standard deviations were calculated by using data from six replicates (optical density) or plate counts (culture) per MOI.
In vivo trials.
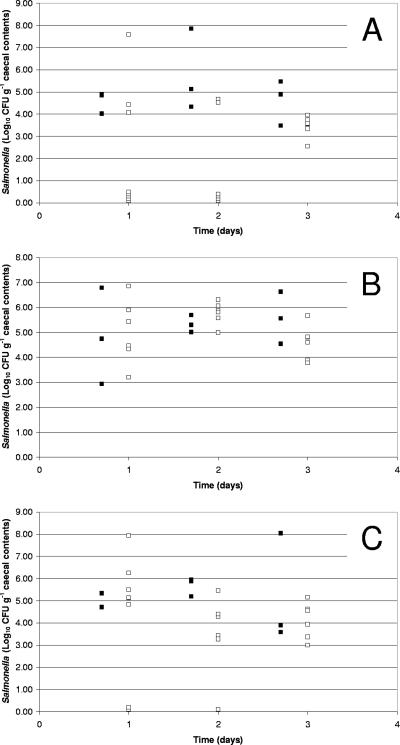
Phages φ151, φ25, and φ10 were used at a lower titer (9.0 log10 PFU ml−1, trial 1) and a higher titer (11.0 log10 PFU ml−1, trial 2) in separate in vivo trials to reduce the numbers of Nalr S. enterica serotype Enteritidis P125109, Hadar 18, and Typhimurium 4/74 bacteria colonizing the ceca of broiler chickens. When the phages were administered at 9.0 log10 PFU, no significant reductions in the cecal carriage of Salmonella in the phage-treated broilers was recorded for any of the serotypes tested over the 6-day duration of the trial. This was despite a significant increase in the titers of phage φ151 (5.2 ± 1.63 log10 PFU g−1 cecal content compared with controls) within 24 h of phage administration (data not shown). The phages were rapidly removed from the chicken intestine in the absence of Salmonella hosts (group B) and were below the limit of detection after 48 h (φ151) or 72 h (φ10, φ25). In the second trial, the chickens were sacrificed over a period of 3 days only, as the results from trial 1 indicated that any decrease in Salmonella numbers following phage treatment should be apparent in this period (data not shown). The results of this second trial are presented in Fig. 3. The higher phage titer corresponded to a significant reduction in the mean cecal colonization by S. enterica serotypes Enteritidis P125109 and Typhimurium 4/74 after 24 h (1.53 ± 2.38 and 3.48 ± 1.88 log10 CFU g−1 cecal content, respectively, compared with the control groups [5.77 ± 1.85 and 5.67 ± 0.41 log10 CFU g−1 cecal content]). These reductions in cecal colonization were significant for both S. enterica serotypes Enteritidis (P < 0.0000001) and Typhimurium (P < 0.000001) by a single-factor ANOVA. No significant differences between control and phage-treated groups were recorded for birds colonized with S. enterica serotype Hadar 18 (5.77 ± 0.45 and 5.34 ± 0.34 log10 CFU g−1 cecal content for phage-treated and control groups, respectively).
FIG. 3.
Efficacy of high-titer phage treatment in reducing Salmonella counts in the ceca of broiler chickens (trial 2). Broiler chickens colonized by S. enterica serotype Enteritidis P125109 (A), Hadar 18 (B), or Typhimurium 4/74 (C) were treated separately with 11.0 log10 PFU of phage φ151, φ25, or φ10, respectively. Salmonella counts from the ceca of individual control birds (▪) and phage-treated birds (□) are presented as log10 CFU g−1 cecal content.
Resistance to phage in vivo.
Following the phage therapy trials, up to six Salmonella colonies were randomly selected from spread plates from the cecal contents of control and phage-treated birds. These colonies were used to prepare lawns onto which phage suspensions were spotted in order to determine the presence of BIMs. A total of 595 colonies were tested in this way (271 for trial 1 and 324 for trial 2). For both trial 1 and trial 2, salmonellas recovered from control animals harbored a subpopulation which was naturally resistant to the phage chosen for therapy. Over the course of trial 1, the number of phage-resistant Salmonella colonies from control animals was 3/24 (12.5%) for S. enterica serotype Enteritidis, 3/23 (13%) for S. enterica serotype Hadar, and 3/44 (6.8%) for S. enterica serotype Typhimurium. Corresponding values for trial 2 were 11/37 (29.7%) for S. enterica serotype Enteritidis, 3/36 (8.3%) for S. enterica serotype Hadar, and 1/52 (1.9%) for S. enterica serotype Typhimurium. Following phage treatment in trial 1, the percentage of BIMs approximately doubled (S. enterica serotype Enteritidis, 11/52 [21.2%]; S. enterica serotype Hadar, 9/39 [23%]; S. enterica serotype Typhimurium, 9/89 [10%]). The percentage of BIMs recovered from phage-treated birds in trial 2 was greater than that in trial 1 for S. enterica serotypes Enteritidis (65/74, 87.8%) and Typhimurium (52/62, 83.9%) but not for serotype Hadar (5/63, 7.93%). The increase in BIMs recovered from phage-treated birds harboring S. enterica serotypes Enteritidis and Typhimurium in trial 2 was significant for both groups (P < 0.001) by the chi-square test. Salmonella colonies recovered from phage-treated birds (n = 20) did not maintain their phage-resistant phenotype when subcultured on BG agar five times. A small-scale trial was performed to establish if phage resistance was maintained in vivo. Three Salmonella BIMs (S. enterica serotypes Enteritidis R1, Hadar R2, and Typhimurium R3) were selected for each serotype from trial 1 and used to reinfect a small group of birds (n = 9) as described above. After 6 days, the birds were sacrificed and the cecal contents were cultured for salmonellas and phage as described above. Colonization levels in the ceca for each resistant Salmonella did not differ appreciably from the controls (data not shown). Additionally, when six colonies from each bird were subsequently screened for phage resistance, the proportion of BIMs did not differ appreciably from the controls.
DISCUSSION
Bacteriophage therapy is undergoing a renaissance in industrialized countries (1). An increasing number of studies have examined the use of phage against food spoilage bacteria or zoonotic pathogens (15, 16). However, for phage therapy to be useful in agricultural applications, it must be tested under conditions which emulate commercial practices. We sought to isolate and use phage to reduce the numbers of three Salmonella serotypes in commercially obtained animals under conditions more congruous with the broiler farm environment. Using specific-pathogen-free animals or applying phage in chick models of Salmonella colonization may produce misleading results as Salmonella colonizes these animals more readily than older birds (10). Applying phage just prior to slaughter is more likely to result in greater efficacy and a reduced proportion of BIMs (8).
Before the three phages were used against Salmonella in vivo, the replication dynamics of each phage-host system were characterized in vitro. Each phage was able to significantly reduce the numbers of their respective Salmonella hosts with MOIs of 10°, 103, and 106. In the case of φ10 applied at an MOI of 106, S. enterica serotype Typhimurium counts were reduced to below the limit of detection over a 24-h period.
It was envisaged that phage treatment would be most efficacious for Salmonella close to the slaughter age of commercially reared broiler chickens (approximately 40 to 42 days in the United Kingdom). Therefore, the efficacy of phage therapy would be maximized by the use of a high titer of bacteriophage to reduce Salmonella colonization by passive inundation (19, 20). In the first trial, the birds were inoculated with 9.0 log10 PFU of phage. This should have resulted in an approximate MOI of 106, which was found to be highly effective in vitro. However, cecal counts of the three Salmonella serotypes were not reduced significantly following phage treatment. The dynamics of phage-bacterium interactions in vivo may be very different from those in vitro because of the viscosity of the gut matrix (29), complex physicochemical environment, and host defenses (8). The numbers of salmonellas in the ceca of commercial broiler chickens and on processed carcasses are generally low (10, 28). This compounds the problems phage have in locating a suitable host in a complex intestinal milieu containing large numbers of “decoy” bacteria and particulate matter. The influence of these decoys may be negligible when the number of target bacteria is high, for example, with Campylobacter (∼7.0 log10 CFU g−1 cecal content). However, as the ratio of target-to-decoy bacteria decreases, the number of phage required to achieve a significant reduction in host numbers increases. Indeed, if the number of target bacteria falls below a minimum number, termed the phage proliferation threshold (19, 30), the large number of phage required may render phage therapy impractical.
In order to assess whether a higher phage MOI would be more effective, a second in vivo trial was performed with 11.0 log10 PFU. It was clear from the second trial that increasing the MOI by 100-fold overcame many of the problems of the low number of host bacteria present in the ceca. On the first day after phage treatment, S. enterica serotype Enteritidis bacterial numbers were reduced significantly by 2.52 log10 CFU g−1. By day 2, the S. enterica serotype Enteritidis count in the phage-treated birds was 1.53 ± 2.38 log10 CFU, compared with 5.77 ± 1.85 log10 CFU in control birds. A significant reduction in S. enterica serotype Typhimurium counts was also recorded for phage-treated birds on day 2 (mean of 3.48 ± 1.88, compared with 5.67 ± 0.41 log10 CFU g−1 for controls). However, no significant reductions in S. enterica serotype Hadar counts were recorded in either trial 1 or trial 2. This was surprising in view of the results obtained for the other serotypes in trial 2 and the activity recorded for φ25 in vitro. The proportion of S. enterica serotype Hadar BIMs recovered from phage-treated birds remained low compared with the other two serotypes (7.9%, compared with 87.8% for S. enterica serotype Enteritidis and 83.9% for S. enterica serotype Typhimurium). If phage therapy relies mainly on passive inundation rather than successive rounds of viral infection and replication, significant numbers of phage are needed to adsorb to individual host cells. S. enterica serotype Hadar 18 may not possess a sufficient number of accessible receptors on the cell surface to allow the adsorption of large numbers of phage. Loss or alteration of the phage receptor(s) or restriction modification systems is unlikely to explain why φ25 was ineffective, as this would have been detected during the in vitro experiments. Nevertheless, it is clear that the behavior of φ25 in vitro was not a reliable indication of its activity in vivo. Identifying the receptor(s) for φ25 may allow more precise bacteriophage therapy through the use of cocktails of phages which adsorb to different receptors. This may also need to be the case for phages which infect S. enterica serotypes Enteritidis and Typhimurium in order to delay the succession of phage-resistant mutants.
This study has demonstrated that bacteriophages can be used to significantly reduce the cecal colonization of S. enterica serotypes Enteritidis and Typhimurium in commercial broiler chickens. Although BIMs were able to colonize chicken ceca within 24 to 48 h of phage treatment, phage resistance was not maintained for long periods either in vitro or in vivo. The results of this study are promising, although further work needs to be undertaken to determine the optimal timing and delivery of bacteriophage in a real-life poultry industry setting.
Acknowledgments
This work was funded by EU FP6 project SUPASALVAC.
We thank Ann Cornish, Pauline Hunt, Charlie Chambers, Maria Rubio, Danilo Hernandez, Tristan Cogan, and Helen Weaver for assistance. We thank Wendy Fielder and Christine Dodd (University of Nottingham) for providing several of the Salmonella strains used in this study. We thank Stefan Hyman and Natalie Allcock (University of Leicester) for expert assistance in electron microscopy.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Anonymous. 2004. Renaissance phage. Nat. Rev. Microbiol. 2:922. [DOI] [PubMed] [Google Scholar]
- 2.Atterbury, R. J. 2006. The age of phage. Poult. Int. 45:18-22. [Google Scholar]
- 3.Atterbury, R. J., P. L. Connerton, C. E. Dodd, C. E. Rees, and I. F. Connerton. 2003. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6302-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dibner, J. J., and J. D. Richards. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634-643. [DOI] [PubMed] [Google Scholar]
- 5.Barrow, P., M. Lovell, and A. Berchieri, Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2005. Salmonella surveillance: annual summary, 2004. Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Cogan, T. A., and T. J. Humphrey. 2003. The rise and fall of Salmonella Enteritidis in the UK. J. Appl. Microbiol. 94(Suppl. 1):114-119. [DOI] [PubMed] [Google Scholar]
- 8.Connerton, P. L., and I. F. Connerton. 2005. Microbial treatments to reduce pathogens in poultry meat, p. 414-427. In G. Mead (ed.), Food safety control in the poultry industry. Woodhead Publishing Ltd., Cambridge.
- 9.Connerton, P. L., C. M. Loc Carrillo, C. Swift, E. Dillon, A. Scott, C. E. Rees, C. E. Dodd, J. Frost, and I. F. Connerton. 2004. Longitudinal study of Campylobacter jejuni bacteriophages and their hosts from broiler chickens. Appl. Environ. Microbiol. 70:3877-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, N. A., L. J. Richardson, J. S. Bailey, D. E. Cosby, J. A. Cason, and M. T. Musgrove. 2005. Bacterial contamination of poultry as a risk to human health, p. 21-35. In G. Mead (ed.), Food safety control in the poultry industry. Woodhead Publishing Ltd., Cambridge, United Kingdom.
- 11.Davies, R. H. 2005. Pathogen populations on poultry farms, p. 101-135. In G. Mead (ed.), Food safety control in the poultry industry. Woodhead Publishing Ltd., Cambridge, United Kingdom.
- 12.El-Shibiny, A., P. L. Connerton, and I. F. Connerton. 2005. Enumeration and diversity of campylobacters and bacteriophages isolated during the rearing cycles of free-range and organic chickens. Appl. Environ. Microbiol. 71:1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Food Safety Authority. 2005. Trends and sources of zoonoses, zoonotic agents and antimicrobial resistance in the European Union in 2004. European Food Safety Authority, Parma, Italy. (Online.) http://www.efsa.europa.eu/en/science/monitoring_zoonoses/reports/1277.html.
- 14.Grajewski, B. A., J. W. Kusek, and H. M. Gelfand. 1985. Development of a bacteriophage typing system for Campylobacter jejuni and Campylobacter coli. J. Clin. Microbiol. 22:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer, G. G. 2005. Bacteriophage control of foodborne bacteria. J. Food Prot. 68:1102-1111. [DOI] [PubMed] [Google Scholar]
- 16.Hudson, J. A., C. Billington, G. Carey-Smith, and G. Greening. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68:426-437. [DOI] [PubMed] [Google Scholar]
- 17.Kutter, E. 1997. Phage therapy: bacteriophages as antibiotics. Evergreen State College, Olympia, WA. (Online.) http://www.evergreen.edu/phage/phagetherapy/phagetherapy.htm.
- 18.Loc Carrillo, C., R. J. Atterbury, A. el-Shibiny, P. L. Connerton, E. Dillon, A. Scott, and I. F. Connerton. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ Microbiol. 71:6554-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne, R. J., and V. A. Jansen. 2001. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208:37-48. [DOI] [PubMed] [Google Scholar]
- 20.Payne, R. J. H., and V. A. Jansen. 2000. Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharm. Ther. 68:225-230. [DOI] [PubMed] [Google Scholar]
- 21.Rohwer, F., and R. Edwards. 2002. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 2, p. 2.64-2.66. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Sklar, I. B., and R. D. Joerger. 2001. Attempts to utilize bacteriophage to combat Salmonella enterica serovar Enteritidis infection in chickens. J. Food Safety 21:15-30. [Google Scholar]
- 24.Smith, H. W., and M. B. Huggins. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129(Pt. 8):2659-2675. [DOI] [PubMed] [Google Scholar]
- 25.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulakvelidze, A., and P. Barrow. 2005. Phage therapy in animals and agribusiness, p. 335-380. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Inc., Boca Raton, FL.
- 27.Summers, W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437-451. [DOI] [PubMed] [Google Scholar]
- 28.Waldroup, A. L., B. M. Rathgeber, and R. H. Forsythe. 1992. Effects of six modifications on the incidence and levels of spoilage and pathogenic organisms on commercial processed postchill broilers. J. Appl. Poult. Res. 2:111-116. [Google Scholar]
- 29.Weld, R. J., C. Butts, and J. A. Heinemann. 2004. Models of phage growth and their applicability to phage therapy. J. Theor. Biol. 227:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Wiggins, B. A., and M. Alexander. 1985. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 49:19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]