Abstract
Endo-β-1,4-xylanases (EC 3.2.1.8; endoxylanases), key enzymes in the degradation of xylan, are considered to play an important role in phytopathogenesis, as they occupy a prominent position in the arsenal of hydrolytic enzymes secreted by phytopathogens to breach the cell wall and invade the plant tissue. Plant endoxylanase inhibitors are increasingly being pinpointed as part of a counterattack mechanism. To understand the surprising XIP-type endoxylanase inhibitor insensitivity of endoxylanases XylA and XylB from the phytopathogen Fusarium graminearum, an extensive mutational study of these enzymes was performed. Using combinatorial and site-directed mutagenesis, the XIP insensitivity of XylA as well as XylB was proven to be solely due to amino acid sequence adaptations in the “thumb” structural region. While XylB residues Cys141, Asp148, and Cys149 were shown to prevent XIP interaction, the XIP insensitivity of XylA could be ascribed to the occurrence of only one aberrant residue, i.e., Val151. This study, in addition to providing a thorough explanation for the XIP insensitivity of both F. graminearum endoxylanases at the molecular level, generated XylA and XylB mutants with altered inhibition specificities and pH optima. As this is the first experimental elucidation of the molecular determinants dictating the specificity of the interaction between endoxylanases of phytopathogenic origin and a plant inhibitor, this work sheds more light on the ongoing evolutionary arms race between plants and phytopathogenic fungi involving recognition of endoxylanases.
The importance of endo-β-1,4-xylanases (EC 3.2.1.8; referred to as endoxylanases) as key enzymes in the degradation of xylan, the predominant hemicellulose in the cell walls of plants and the second most abundant polysaccharide on earth, cannot be overestimated. They are produced by a large variety of organisms including bacteria, fungi, and plants and are of significant importance in many physiological, pathological, and even biotechnological processes which involve degradation or remodeling of the plant cell wall. The physiological role of plant endogenous endoxylanases is linked to their involvement in processes such as secondary cell wall biogenesis and metabolism (1, 36), germination (9, 41), and the initiation of sexual reproduction by facilitating pollen tube penetration (35). Endoxylanases secreted by phytopathogenic microorganisms are generally considered (4) and occasionally proven (7) to be essential components of their offensive arsenal to penetrate and colonize plant tissues. Apart from this, endoxylanases of microbial origin are increasingly utilized by mankind, as they have a high impact on biotechnological applications in environment-related technologies such as biological bleaching of chemical pulps (2), bioconversion of agricultural residues to fuel ethanol (33), and industrial feed (8) and food processing (10).
An important factor governing endoxylanase functionality is formed by plant endoxylanase inhibitors, which are able to reduce or completely block endoxylanolytic activity. While, in recent years, much research effort has been directed towards evaluating and manipulating the influence of these inhibitors for applications of endoxylanases in biotechnological processes (11, 18, 27, 32, 40), little if anything is known about their role in plant physiology and defense. Their relevance in these contexts, though, should not be underestimated as both types of endoxylanase inhibitors described to date, i.e., Triticum aestivum xylanase inhibitor (TAXI) type (19) and xylanase inhibitor protein (XIP) type (26), are wound and pathogen inducible (23, 24) and occur as multiple-isoform families widely represented within the plant kingdom (13, 20, 31). It remains to be demonstrated whether the recently described TLXI inhibitors (15) are also wound and/or pathogen inducible. To obtain comprehensive insight into the role of endoxylanases and their inhibitors in plant-microbe interactions, knowledge of the specific relationships between both interacting proteins is crucial. The currently known TAXI-type inhibitors inhibit all thus far tested microbial endoxylanases of glycosyl hydrolase family 11 (GH11), but not those of GH10 (19). XIP-type inhibitors, on the other hand, typically inhibit GH10 and GH11 endoxylanases from fungal sources (26). Surprisingly however, two GH11 endoxylanases (XylA and XylB) from the phytopathogenic fungus Fusarium graminearum were found to be insensitive to XIP-I (5). From the crystal structures of the TAXI-I-Aspergillus niger ExlA (34) and XIP-I-Penicillium funiculosum XynC endoxylanase (30) complexes, it is clear that the inhibition strategy is based on substrate-mimicking contacts. Inhibitor insensitivity must thus depend on details in the architecture of the areas delineating the substrate binding groove, which disable interactions with the inhibitor while leaving those with the substrate unaffected. Consequently, our understanding of the biological significance of endoxylanase-inhibitor interactions would seriously benefit from a thorough analysis of the contribution of individual amino acids involved in inhibitor-mediated recognition of endoxylanases, in particular those of phytopathogenic origin. To this aim, we here report an extensive mutational study of both F. graminearum GH11 endoxylanases, based on a combination of three-dimensional modeling and the use of a previously developed phage display selection system (3).
MATERIALS AND METHODS
Strains, plasmids, phagemid, and helper phage.
Escherichia coli XL1-Blue MRF′ (Stratagene, La Jolla, CA) was used as the host strain for subcloning. E. coli BL21(DE3) was used as the host strain for heterologous expression via expression vector pQE-EN. E. coli CJ236 (New England Biolabs, Beverly, MA) and E. coli BMH71-18 mutS (TaKaRa, Shuzo Co., Shiga, Japan) were used for preparation of deoxyuridine-containing single-stranded DNA and for transformation of the phagemid pHOS31 library, respectively. E. coli TG1 was used as the acceptor strain for helper phage VCSM13 (Stratagene) and phage infections during successive rounds of biopanning.
Homology modeling of XylA and XylB.
Three-dimensional models of XylA and XylB were built with the aid of the automated comparative protein modeling server SWISS-MODEL (21) on the basis of solved crystal structures of GH11 endoxylanase homologues (PDB accession codes: 1m4wA, 1pvxA, 1h1aA, 1h1aB, and 1xypA for XylA and 1xyoA, 1reeA, 1xypA, 1enxB, and 1refA for XylB). The structures were validated by using the VERIFY-3D (14) and ANOLEA (28) programs.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) with either pQE-EN-xylA or pQE-EN-xylB (5) as the template DNA and a pair of complementary mutagenic primers according to the manufacturer's instructions. Sequences of the forward primers for each mutation are indicated in Table 1.
TABLE 1.
Oligonucleotides used for mutagenesis
| Target residue(s) | Sequence (5′-)a | Substitution(s) |
|---|---|---|
| XylA N65 | GGCGCAACACTGGTGACTTTGTCGGTGG | D65 |
| XylA Q144 | GTCCACCCGTTACAACCAGCCTTCGATC | N144 |
| XylA V151 | CTTCGATCGACGGTACTCAGACCTTCAACC | T151 |
| XylB N62 | GGCAACCGTGGTGACCACGTCGGTGG | D62 |
| XylB library | ||
| T38P39S40A41 | GTTGCCGTTGGTGTAGGTGAC(A/C)(G/T)CGC(T/C)G(G/C)(C/G)G(G/C)(T/C)ATCGGTCCAGAAGGAGAAG | T38, G38, S38, A38; P39, G39, R39, A39; S40, G40; A41, D41, E41 |
| N62H63 | CCTTACCACCGACG(T/A)(G/A)GT(T/C)ACCAC GGTTGCCGTTC | N62, D62; H63, F63, L63, Y63 |
| C141-D148C149 | CTGCTGGAAGGTCTTG(C/G)(A/T)ACCCTCGATGGAAGGAGCA(C/T)(A/T)GTTGCGAGTGCTG | C141, N141, Y141, S141; C149, T149, S149 |
Degeneracies are indicated by parentheses, underlined nucleotides correspond to mutated codons, and boldface nucleotides correspond to mutations.
Construction of the xylB combinatorial library.
Phagemid pHOS31-xylB was constructed for use as the template for library construction. pHOS31-xylB is comparable to previously described phagemids (3) designed to display GH11 endoxylanases on the surface of M13 bacteriophage as C-terminal fusions to phage minor coat protein g3p. pQE-EN-xylB was digested with BglII to obtain the xylB insert for subcloning as a BglII fragment in pHOS31. The phage-displayed combinatorial xylB library was constructed essentially as described previously (6). Briefly, pHOS31-xylB deoxyuridine-containing single-stranded DNA was used as the template for the Kunkel mutagenesis method with mutagenic oligonucleotides (Table 1) designed to introduce mutations at the desired sites. The mutagenesis reaction mixture was electroporated into E. coli BMH71-18 mutS. The library obtained after several transformations contained ∼2 × 109 unique members, and DNA sequencing of several randomly picked clones of the naive library revealed that 50% of these had incorporated all three mutagenic oligonucleotides. Hence, the theoretical diversity for combinatorial mutagenesis at the nine selected positions (∼1.6 × 104) was exceeded by approximately 105-fold.
Biopanning of the xylB combinatorial library.
Maxisorb plate (Nalge Nunc International, Naperville, IL) wells were coated with ∼1 μg endoxylanase inhibitor XIP-I or TAXI-I, and ∼4 × 1010 CFU library phages was added. Following a 2-hour incubation to allow phage binding, the plates were washed six times with phosphate-buffered saline (PBS) and six times with PBS containing 0.1% Tween 20. Bound phages were eluted by a 10-minute incubation with 100 mM triethylamine (pH 12.0), and the eluate was neutralized with 1.0 M Tris-HCl, pH 7.4. For all selections, an equal number of uncoated wells were used as a negative control. A fraction (∼1/100 volume) of the neutralized eluted phage solution was plated in serial dilutions on selective agar plates to determine the phage titers. Eluted phages were amplified in E. coli TG1 and used for further rounds of selection. The selection process was monitored by titering the phage suspensions before selection and after elution.
DNA sequencing.
Phagemid and expression plasmid inserts were sequenced on an ABI 377 sequencer using ABI PRISM BigDye Terminator chemistry (Applied Biosystems, Foster City, CA). The Sequencher 4.1 package for Apple Macintosh (Gene Codes, Ann Arbor, MI) was used to edit and align obtained DNA sequences.
Recombinant expression and purification of endoxylanases and mutants.
F. graminearum endoxylanases XylA and XylB and mutants thereof were heterologously expressed using vector pQE-EN under previously derived optimal expression conditions (5). Ni-nitrilotriacetic acid magnetic agarose beads (QIAGEN GmbH, Düsseldorf, Germany) were used to purify the His6-tagged proteins from the lysate under native conditions.
Endoxylanase activity and inhibition assay.
Endoxylanase activity and inhibition activities were determined with the colorimetric Xylazyme-AX tablet method (Megazyme, Bray, Ireland) and a variant thereof, respectively, as described before (5). Xylazyme-AX tablets consist of azurine-cross-linked wheat arabinoxylan, which, upon hydrolysis by endoxylanases, produces water-soluble dyed fragments. In the assay, the rate of release (increase of absorbance at 590 nm [A590]) can be related directly to enzyme activity. One enzyme unit (EU) corresponds to an increase in A590 of 1.0 under the conditions in the assay. The pH for maximal endoxylanase activity was determined by measuring activities at various pHs. To this end, Ni-nitrilotriacetic acid-purified endoxylanase solutions were prepared in sodium acetate (250 mM, pH 3.0, 4.0, 5.0, and 6.0) and sodium phosphate buffer (250 mM, pH 6.0, 7.0, 8.0, and 9.0). The inhibition assay is identical to that described above, except that endoxylanase solutions (approximately 1 EU in 250 μl) were preincubated for 30 min with an equal amount of inhibitor solution at room temperature before addition of a Xylazyme-AX tablet. Inhibition assays were performed with ∼1 μg recombinant XIP-I or TAXI-I, obtained as described earlier (3, 16). The statistical significance of differences was tested at a significance level (P) of <0.05 using a one-tailed Student t test.
RESULTS AND DISCUSSION
Molecular modeling of XylA and XylB.
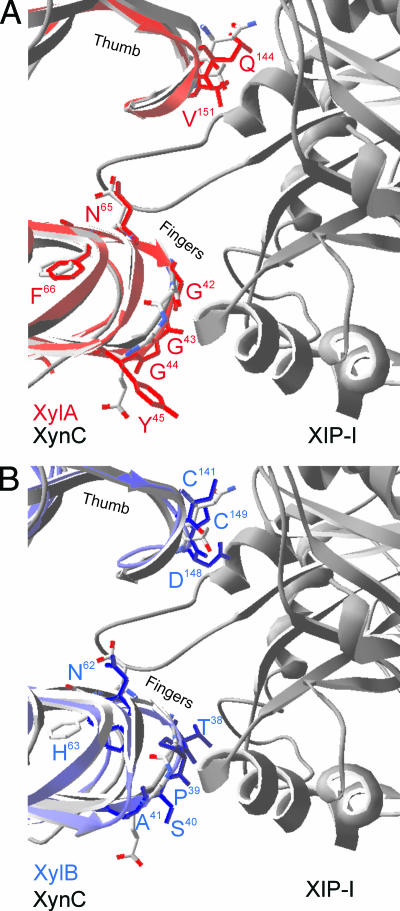
A thorough analysis of the contribution of the single amino acids in the GH11 endoxylanase-XIP interactions is difficult, due to the high number of contacts established in PDB entry 1TE1, to date the only solved crystal structure of XIP in complex with a GH11 endoxylanase (i.e., XynC from P. funiculosum) (30). Hence, in order to obtain comprehensive insight into the XIP insensitivity of both F. graminearum GH11 endoxylanases, we performed an extensive mutational analysis using homology models of both endoxylanases as starting point. As for other GH11 endoxylanases, the generated three-dimensional models of XylA and XylB adopt a β-jelly roll fold, in which two large β-pleated sheets and one α-helix form a structure that resembles a partly closed right hand. A two-β-strand “thumb” forms a lid over the active site, which is located in the “palm” (39). The homology models were shown to be reliable by their Ramachandran plots and VERIFY-3D (14) and ANOLEA (28) analyses (data not shown). Furthermore, superimposition of the XylA and XylB model on the P. funiculosum XynC endoxylanase structure resulted in pairwise root-mean-square values of 0.84 Å and 1.02 Å, respectively. As shown in Fig. 1A, XylA does not exhibit significant structural differences compared to XynC, either in the thumb region or in any other XIP-I contact region. Consequently, as previously predicted from sequence comparisons (5), the structural features responsible for the XIP insensitivity of XylA can be restricted to one or both of the following amino acid differences: Asn124XynC→Gln144XylA and Thr131XynC→Val151XylA. A different situation occurs for XylB, however. Especially the hairpin loop region of the XylB “thumb” is predicted to adopt a strikingly distinctive conformation that would introduce steric clashes with XIP-I, as envisaged by the structural alignment of the model and the superimposed solved XynC-XIP-I complex (Fig. 1B). In addition, at the “finger turns” several other considerable structural dissimilarities in XIP-I contact regions of XylB, particularly including residues 38TPSA41 and 62NH63, are revealed. Interestingly, the asparagine residue of the latter finger turn region is located in the active site at the hydrogen-bonding distance of the acid/base catalyst. At the corresponding position of the A. niger GH11 endoxylanase (ExlA) is found an aspartic acid, which was shown to be critical for XIP-I binding (38). As also XylA exhibits an asparagine (Asn65) at this key position, we wondered whether replacement of these asparagines by aspartic acids would have any influence on the insensitivity of both F. graminearum GH11 endoxylanases to XIP-I.
FIG. 1.
Molecular models of F. graminearum GH11 endoxylanases XylA (A) and XylB (B) superimposed upon the solved cocrystal structure of P. funiculosum XynC in complex with XIP-I. Ribbon representations of XynC (gray, left side), XIP-I (gray, right side), XylA (red), and XylB (blue) are shown. The “thumb” and “finger” regions, as well as side chains of putative specificity-determining residues, are indicated.
Inhibitor sensitivity of site-specific XylA mutants.
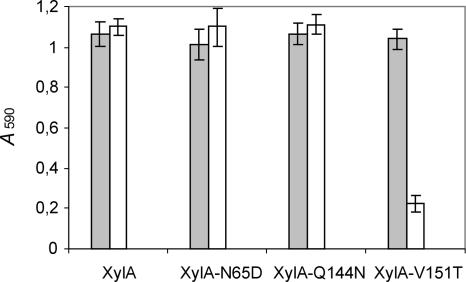
Since, according to the above-described modeling analysis, the XIP insensitivity of XylA could be attributed to only two amino acid residues, Gln144 and Val151, site-directed mutants XylA-Q144N and XylA-V151T were generated and tested for inhibition sensitivity. In addition, a site-directed mutant in which the asparagine at the position adjacent to the general acid/base catalytic residue was replaced by an aspartic acid (XylA-N65D) was created. Figure 2 demonstrates that neither replacement of Gln at position 144 of XylA by an Asn nor replacement of Asn at position 65 by Asp affects the absence of XIP-I-mediated inhibition. The XylA-V151T mutant, on the other hand, is strongly inhibited by XIP-I (P = 1.1 × 10−5). Hence, based on these data, the insensitivity of XylA to XIP-I is exclusively due to the valine residue occurring at position 151. This can be explained by the fact that this Val side chain, unlike the Thr side chain, is unable to form a hydrogen bond with Asn147 of XIP-I, but instead collides with the latter residue. The Gln at position 144 of XylA turns out not to be responsible for XIP-I insensitivity, although a highly conserved Asn at the corresponding position of the A. niger GH11 endoxylanase (ExlA) was shown to be crucial for XIP-I interaction (37). However, even though different amino acid residues at different positions are involved, a common property of XIP-I binding to XylA and ExlA is that molecular recognition of both enzymes by XIP-I depends on subtle sequence differences and hence can be altered by a single amino acid substitution. The same applies for XynII from Trichoderma longibrachiatum, which loses its sensitivity to XIP-I upon mutagenesis of Thr at position 131 to Arg (6). While the latter mutant was retrieved by in vitro selection, the molecular architecture of the thumb region of XylA appears to be specifically adapted during evolution in order to escape inhibition by plant inhibitors, thereby maintaining the capacity of F. graminearum to breach the plant cell wall with the wealthy arsenal of xylan-degrading enzymes secreted (22).
FIG. 2.
Endoxylanase inhibition assay of XylA mutants with altered inhibition characteristics. Endoxylanase activities in the absence (gray bars) and presence (white bars) of XIP-I are shown. Means and standard deviations of triplicate experiments are shown.
Although the inhibition specificity of the XylA-N65D mutant was unaltered (Fig. 2), its pH optimum was found to be lowered by approximately 1 pH unit (data not shown). Consequently, Asn65 turns out to be a key residue in determining the relatively high pH value (pH ∼8) for optimal activity of XylA. These data are in line with the general observed correlation between the presence of an asparagine or an aspartic acid residue at the position adjacent to the acid/base catalyst and the “alkaline” or “acidic” pH optima of GH11 endoxylanases, respectively (25).
Screening of a phage-displayed XylB combinatorial library against endoxylanase inhibitors.
In the case of XylB, the involvement of additional regions outside the thumb region in conferring insensitivity to XIP-I could not be excluded on basis of the modeling analysis. Therefore, we opted for a combinatorial mutational approach, which allowed us to experimentally verify the role of multiple amino acid residues located in the “thumb” region as well as the above-mentioned “finger turn” regions simultaneously. By means of three degenerate primers (Table 1), a phage-displayed combinatorial mutant library of XylB was constructed in which the nine residues at positions 38, 39, 40, 41, 62, 63, 141, 148, and 149 were preferentially allowed to be either the wild-type residues or the corresponding residues of endoxylanases sensitive to endoxylanase inhibitor XIP-I (Table 1). The phage-displayed combinatorial mutant library was subjected to three consecutive rounds of biopanning against XIP-I and TAXI-I. The titer of phages selected against TAXI-I after the first round was already considerably higher than that of phages selected in control wells (uncoated wells blocked with bovine serum albumin) (data not shown). This initial strong enrichment was almost unchanged throughout the panning procedure, reflecting the fact that few if any substitutions affecting TAXI-I binding were included in the library. In contrast, the enrichment ratio of phages selected against XIP-I increased 10-fold after each panning round. This clearly pointed to functional selection of XIP-I binding phages.
After three rounds, the XylB sequences of several clones enriched via biopanning on XIP-I and TAXI-I were determined. As expected, no notable mutations were present in the clones enriched against TAXI-I (data not shown). This is in contrast to clones enriched against XIP-I, for which obvious sequence preferences were obtained. Besides deletion of Asp at position 148, the enriched mutants typically displayed the following mutations: Cys141→Asn141 and Cys149→Thr149. Based on these data, it can be deduced that these thumb residues play a crucial role in preventing XIP-I-mediated inhibition of XylB. As the wild-type 62NH63 sequence is well represented among the enriched clones, this finger turn region is most likely not responsible for the insensitivity of XylB to XIP-I. The other finger turn region in close proximity of XIP-I (38TPSA41), however, was clearly adapted during the selection process, as this region diverged to 38GGGD41 in the majority of the enriched clones.
Inhibitor-sensitive combinatorial and site-directed XylB mutants.
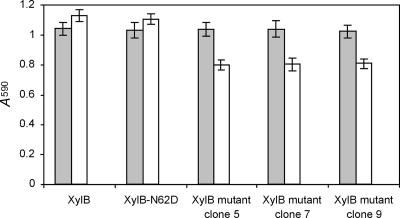
In order to experimentally confirm the above-outlined findings, three XylB mutants selected via biopanning (clones 5, 7, and 9) were subcloned in expression vector pQE-EN and recombinantly expressed. Clone 5 (XylB-38TPSA41-62NH63-141N-T149) was chosen because its amino acid sequence adaptation is restricted to the thumb region. Clone 7 (XylB-38GGGD41-62NH63-141N-T149), on the other hand, displays on each mutated position the amino acid residue which was most frequently represented in the enriched clones. This XylB mutant was identical to clone 9, except for Phe at position 63. Hence, comparison of the XIP-I-mediated inhibition of both mutants would allow us to determine the effect of the latter residue on the XIP-I interaction. In line with expectations, the enzymatic activities of the three selected XylB mutants were also significantly lowered upon preincubation with XIP-I (P of 9.4 × 10−4, 1.9 × 10−3, and 1.1 × 10−3 for XylB mutant clone 5, clone 7, and clone 9, respectively) (Fig. 3). The mutations thus generate inhibition sensitivity, but to a lesser extent than the XylA-V151T mutant (Fig. 2). In other words, the created interactions with XIP-I do not lead to strong inhibition, indicating that the complexes formed are rather weak. Regarding XIP insensitivity, it can be deduced that only the thumb residues play a critical role, since the degree of inhibition of clone 5 (harboring mutations exclusively at the thumb region) was comparable to that of clones 7 and 9 (harboring additional mutations outside the thumb region). Apparently, the slightly different conformation of the thumb and the loss of hydrogen bonds with Gly179, Ala214, and Asn147 of XIP-I (30) due to aberrant sequence differences Asn124XynC→Cys141XylB and Thr131XynC→Cys149XylB weaken the interaction to a sufficient extent to prevent XIP-I-mediated inhibition of XylB. Remarkably, the presence of both cysteine residues in the thumb region of this F. graminearum endoxylanase is a unique feature, as examination of the Pfam alignment (17) (accession number PF00457) revealed that none of the other 253 GH11 endoxylanase sequences known to date display Cys residues in their thumb regions. Of particular interest in this context is the recent generation of a Thermobacillus xylanilyticus GH11 endoxylanase (XylTx) mutant in which Pro114, Ser115, and Ile116, which form the tip of the thumb, were replaced by a new triplet harboring a cysteine residue (Pro114-Gly115-Cys116) (29). Intriguingly, this unnatural triplet was found to confer an increase in enzymatic activity. It has been suggested that this was due to the formation of a weak hydrogen bond between the sulfhydryl group of Cys and hydroxyls of the xylose residue present in the catalytic groove. Whether or not the cysteine residues in the thumb region of XylB have a dual role, that is, improving substrate binding in addition to preventing inhibitor interaction, is not clear at this moment and will need to be further investigated.
FIG. 3.
Endoxylanase inhibition assay of XylB mutants with altered inhibition characteristics. Endoxylanase activities in the absence (gray bars) and presence (white bars) of XIP-I are shown. Clones 5, 7, and 9 represent the XylB-38TPSA41-62NH63-141N-T149, XylB-38GGGD41-62NH63-141N-T149, and XylB-38GGGD41-62NF63-141N-T149 mutants, respectively. Means and standard deviations of triplicate experiments are shown.
Since Asn at position 62 was found in all enriched sequenced clones, we also tested the inhibition sensitivity of an XylB-N62D site-directed mutant. However, as for XylA-N65D, the mutant was still insensitive to XIP-I (Fig. 3). The pH optimum of the XylB-N62D variant was again lowered by approximately 1 pH unit (data not shown). Hence, also in the case of XylB, the asparagine residue adjacent to the acid/base catalyst plays a pivotal role in fine-tuning the pH for maximal enzymatic activity to neutral (pH ∼7).
Comparative analysis of the thumb sequences of GH11 endoxylanases from phytopathogenic fungi.
It is intriguing how subtle amino acid variations lie at the basis of the XIP-I insensitivity of both F. graminearum endoxylanases. In the case of XylA, this sequence diversity could even be restricted to the Val side chain occurring at position 151. These findings, in all probability, reflect the highly specific adaptations of these enzymes during evolution in order to evade inhibition by inhibitors of plant origin. As the XIP-I specificity-determining residues of both F. graminearum endoxylanases were found to be restricted to the thumb region, a comparative analysis of the thumb regions of other available GH11 endoxylanase sequences from phytopathogenic fungi was performed (Table 2). Based on this sequence analysis, Mycosphaerella graminicola and Phaeosphaeria nodorum are predicted to produce at least one putatively XIP-insensitive endoxylanase. Indeed, the thumb region of M. graminicola endoxylanase M.gra1 harbors an Arg residue, which most likely clashes with the inhibitor instead of forming a hydrogen bond with it, similar to the Val at the corresponding position of XylA. P. nodorum endoxylanase P.nod6, on the other hand, exhibits an insertion of an Asp comparable to the thumb sequence of XylB. Further aberrant thumb sequences are noticed in Botryotinia fuckeliana endoxylanase B.fuc2, Sclerotinia sclerotiorum endoxylanase S.scl1, and Nectria haematococca endoxylanase N.hae3, as none of these display the highly conserved Pro-Ser-Ile triplet (in the last, even two additional residues are lacking, causing a strikingly reduced thumb region).
TABLE 2.
Comparative analysis of the thumb regions of GH11 endoxylanase sequences from other phytopathogenic fungi
| Endoxylanasea | Data bank referenceb | Thumb sequencec |
|---|---|---|
| A.bra1 | WUSTL/Cont6.39 | NQPSIDG-TATF |
| A.bra2 | WUSTL/Cont0.168 | NQPSIDG-TKTF |
| A.bra3 | WUSTL/Cont6.49 | NKPSIEG-TRTF |
| A.bra4 | WUSTL/Cont4.272 | NQPSIQG-TTTF |
| A.sp | DQ641271 | NQPSIDG-TATF |
| A.pis | Z68891 | NQPSIDG-TQTF |
| B.rho | CS272686 | NAPSIDG-TQTF |
| B.fuc1 | AAID01000748 | NQPSIIG-TATF |
| B.fuc2 | AAID01000419 | NQPSVQG-TATF |
| B.fuc3 | AAID01003217 | NEPSIVG-TATF |
| C.pur | Y16969 | NQPSILG-TNTF |
| C.car1 | L13596 | NQPSIDG-TRTF |
| C.car2 | U58915 | NQPSIDG-TRTF |
| C.car3 | U58916 | NQPSIVG-TTTF |
| C.sat | AJ004802 | NQPSIDG-TRTF |
| F.oxy1 | AF246831 | QQPSIDG-TQTF |
| F.oxy2 | AF246830 | NAPSIDG-TQTF |
| G.mon1 | AAIM02000174 | NQPSIDG-QQTF |
| G.mon2 | AAIM02000091 | NAPSIDG-TQTF |
| G.mon3 | AAIM02000113 | NAPSIEG-NKTF |
| M.gri1 | AACU02000386 | NQPSIEG-TRTF |
| M.gri2 | AACU02000365 | NQPSIDG-TRTF |
| M.gri3 | AACU02000636 | NQPSIDG-TKTF |
| M.gri4 | AACU02000633 | NKPSIEG-TRTF |
| M.gri5 | AACU02000500 | NAPSIIG-DADF |
| M.gra1 | Cogeme/mg[0935] | NQPSIDA-RRPF |
| M.gra2 | JGI/Mycgr/scaf4:1008265:1008944 | NQPSIDG-TQTF |
| N.hae1 | JGI/Necha2/34530 | NAPSIEG-TQTF |
| N.hae2 | JGI/Necha2/106153 | NAPSIEG-TRTF |
| N.hae3 | JGI/Necha2/19304 | NVSSSE---QTV |
| P.nod1 | AAGI01000353 | NQPSIDG-TRTF |
| P.nod2 | AAGI01000307 | NQPSIDG-TSTF |
| P.nod3 | AAGI01000275 | NQPSIDG-TSTF |
| P.nod4 | AAGI01000203 | NQPSIQG-TSTF |
| P.nod5 | AAGI01000226 | QKPSIEG-TRTF |
| P.nod6 | AAGI01000183 | NAPSIIGDNTDF |
| S.com | AAS31739 | NAPSIDG-TQTF |
| S.scl1 | AAGT01000462 | NQPSVQG-TATF |
| S.scl2 | AAGT01000343 | NEPSIVG-TATF |
| S.scl3 | AAGT01000133 | NEPSIVG-TATF |
| S.tur1 | AJ238895 | NQPSIDG-TRTF |
| S.tur2 | AJ548879 | NQPSIDG-TRTF |
| U.may | AACP01000245 | DQPSIQG-TKTF |
| V.dah | DQ449069 | NQPSIDG-TRTF |
Putative endoxylanase genes were identified by a tblastn search with the XylA and XylB sequences in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/), FGI (http://www.broad.mit.edu/annotation/fgi/), JGI (http://www.jgi.doe.gov/), Cogeme (http://www.cogeme.man.ac.uk/), TIGR (http://www.tigr.org/), Trust Sanger Center (http://www.sanger.ac.uk/), and WUSTL (http://genome.wustl.edu/). Derivations of endoxylanase designations not introduced in the text are as follows: A.bra, Alternaria brassicicola; A.sp, Alternaria sp. strain HB186; A.pis, Ascochyta pisi; B.rho, Botryosphaeria rhodina (Lasiodiplodia theobromae); C.pur, Claviceps purpurea (Sphacelia purpurea); C.car, Cochliobolus carbonum (Bipolaris zeicola); C.sat, Cochliobolus sativus (Bipolaris sorokiniana); F.oxy, Fusarium oxysporum; G.mon, Gibberella moniliformis (Fusarium verticillioides); M.gri, Magnaporthe grisea; S.com, Schizophyllum commune; S.tur, Setosphaeria turcica (Helminthosporium turcicum); U.may, Ustilago maydis; V.dah, Verticillium dahliae.
With the exception of the codes that are preceded by the above-mentioned genome centers (WUSTL, JGI, and Cogeme), the data bank references refer to GenBank accession numbers.
Aberrant sequence characteristics are underlined.
This work, as the first study shedding light upon the basis of the molecular recognition properties of a plant inhibitor and endoxylanases from a phytopathogenic source, indicates a more specific interference with key enzymes in plant carbohydrate degradation upon pathogen attack. In addition, the obtained insights contribute to our understanding of the outcome of usage of microbial endoxylanases in biotechnological applications. Of particular interest in this context is the recent finding that feeding Fusarium-infected wheat positively influences broiler performance and nutritional physiology, which is explained by the fact that Fusarium infection-related increases in endoxylanase activities compensate for the deleterious effects of the fungal-origin mycotoxins (12).
Acknowledgments
This work was financially supported by The Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (Brussels, Belgium) (scholarship and postdoctoral research fellowship for T. Beliën), Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Brussels, Belgium), and Bijzonder Onderzoeksfonds (K. U. Leuven, Belgium) (postdoctoral research fellowships for S. Van Campenhout).
Footnotes
Published ahead of print on 18 May 2007.
REFERENCES
- 1.Aspeborg, H., J. Schrader, P. M. Coutinho, M. Stam, A. Kallas, S. Djerbi, P. Nilsson, S. Denman, B. Amini, F. Sterky, E. Master, G. Sandberg, E. Mellerowicz, B. Sundberg, B. Henrissat, and T. T. Teeri. 2005. Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137:983-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajpai, P. 2004. Biological bleaching of chemical pulps. Crit. Rev. Biotechnol. 24:1-58. [DOI] [PubMed] [Google Scholar]
- 3.Beliën, T., K. Hertveldt, K. Van den Brande, J. Robben, S. Van Campenhout, and G. Volckaert. 2005. Functional display of family 11 endoxylanases on the surface of phage M13. J. Biotechnol. 115:249-260. [DOI] [PubMed] [Google Scholar]
- 4.Beliën, T., S. Van Campenhout, J. Robben, and G. Volckaert. 2006. Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant-Microbe Interact. 19:1072-1081. [DOI] [PubMed] [Google Scholar]
- 5.Beliën, T., S. Van Campenhout, M. Van Acker, and G. Volckaert. 2005. Cloning and characterization of two endoxylanases from the cereal phytopathogen Fusarium graminearum and their inhibition profile against endoxylanase inhibitors from wheat. Biochem. Biophys. Res. Commun. 327:407-414. [DOI] [PubMed] [Google Scholar]
- 6.Beliën, T., S. Van Campenhout, A. Vanden Bosch, T. M. Bourgois, S. Rombouts, J. Robben, C. M. Courtin, J. A. Delcour, and G. Volckaert. 2007. Engineering molecular recognition of endoxylanase enzymes and their inhibitors through phage display. J. Mol. Recognit. 20:103-112. [DOI] [PubMed] [Google Scholar]
- 7.Brito, N., J. J. Espino, and C. Gonzalez. 2006. The endo-beta-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol. Plant-Microbe Interact. 19:25-32. [DOI] [PubMed] [Google Scholar]
- 8.Brufau, J., M. Francesch, and A. M. Perez-Vendrell. 2006. The use of enzymes to improve cereal diets for animal feeding. J. Sci. Food Agric. 86:1705-1713. [Google Scholar]
- 9.Caspers, M. P., F. Lok, K. M. C. Sinjorgo, M. J. van Zeijl, K. A. Nielsen, and V. Cameron-Mills. 2001. Synthesis, processing and export of cytoplasmic endo-β-1,4-xylanase from barley aleurone during germination. Plant J. 26:191-204. [DOI] [PubMed] [Google Scholar]
- 10.Courtin, C. M., and J. A. Delcour. 2002. Arabinoxylans and endoxylanases in wheat flour bread-making. J. Cereal Sci. 35:225-243. [Google Scholar]
- 11.Courtin, C. M., W. Gys, K. Gebruers, and J. A. Delcour. 2005. Evidence for the involvement of arabinoxylan and xylanases in refrigerated dough syruping. J. Agric. Food Chem. 53:7623-7629. [DOI] [PubMed] [Google Scholar]
- 12.Dänicke, S., H. Valenta, and S. Matthes. 2007. On the interactions between Fusarium toxin-contaminated wheat and nonstarch polysaccharide hydrolyzing enzymes in diets of broilers on performance, intestinal viscosity, and carryover of deoxynivalenol. Poult. Sci. 86:291-298. [DOI] [PubMed] [Google Scholar]
- 13.Durand, A., R. Hughes, A. Roussel, R. Flatman, B. Henrissat, and N. Juge. 2005. Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J. 272:1745-1755. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg, D., R. Luthy, and J. U. Bowie. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 277:396-404. [DOI] [PubMed] [Google Scholar]
- 15.Fierens, E., S. Rombouts, K. Gebruers, H. Goesaert, K. Brijs, J. Beaugrand, G. Volckaert, S. Van Campenhout, P. Proost, C. M. Courtin, and J. A. Delcour. 2007. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem. J. 403:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierens, K., N. Geudens, K. Brijs, C. M. Courtin, K. Gebruers, J. Robben, S. Van Campenhout, G. Volckaert, and J. A. Delcour. 2004. High-level expression, purification, and characterization of recombinant wheat xylanase inhibitor TAXI-I secreted by the yeast Pichia pastoris. Protein Expr. Purif. 37:39-46. [DOI] [PubMed] [Google Scholar]
- 17.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frederix, S. A., C. M. Courtin, and J. A. Delcour. 2004. Substrate selectivity and inhibitor sensitivity affect xylanase functionality in wheat flour gluten-starch separation. J. Cereal Sci. 40:41-49. [Google Scholar]
- 19.Gebruers, K., K. Brijs, C. M. Courtin, H. Goesaert, G. Raedschelders, J. Robben, J. F. Sørensen, S. Van Campenhout, and J. A. Delcour. 2004. Properties of TAXI-type endoxylanase inhibitors. Biochim. Biophys. Acta 1696:213-221. [DOI] [PubMed] [Google Scholar]
- 20.Goesaert, H., G. Elliott, P. A. Kroon, K. Gebruers, C. M. Courtin, J. Robben, J. A. Delcour, and N. Juge. 2004. Occurrence of proteinaceous endoxylanase inhibitors in cereals. Biochim. Biophys. Acta 1696:193-202. [DOI] [PubMed] [Google Scholar]
- 21.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 22.Hatsch, D., V. Phalip, E. Petkovski, and J. M. Jeltsch. 2006. Fusarium graminearum on plant cell wall: no fewer than 30 xylanase genes transcribed. Biochem. Biophys. Res. Commun. 345:959-966. [DOI] [PubMed] [Google Scholar]
- 23.Igawa, T., T. Ochiai-Fukuda, N. Takahashi-Ando, S. Ohsato, T. Shibata, I. Yamaguchi, and M. Kimura. 2004. New TAXI-type xylanase inhibitor genes are inducible by pathogens and wounding in hexaploid wheat. Plant Cell Physiol. 45:1347-1360. [DOI] [PubMed] [Google Scholar]
- 24.Igawa, T., T. Tokai, T. Kudo, I. Yamaguchi, and M. Kimura. 2005. A wheat xylanase inhibitor gene, Xip-I, but not Taxi-I, is significantly induced by biotic and abiotic signals that trigger plant defense. Biosci. Biotechnol. Biochem. 69:1058-1063. [DOI] [PubMed] [Google Scholar]
- 25.Joshi, M. D., G. Sidhu, I. Pot, G. D. Brayer, S. G. Withers, and L. P. McIntosh. 2000. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 299:255-279. [DOI] [PubMed] [Google Scholar]
- 26.Juge, N., F. Payan, and G. Williamson. 2004. XIP-I, a xylanase inhibitor protein from wheat: a novel protein function. Biochim. Biophys. Acta 1696:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Juge, N., and B. Svensson. 2006. Proteinaceous inhibitors of carbohydrate-active enzymes in cereals: implication in agriculture, cereal processing and nutrition. J. Sci. Food Agric. 86:1573-1586. [Google Scholar]
- 28.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 29.Paes, G., V. Tran, M. Takahashi, I. Boukari, and M. J. O'Donohue. 2007. New insights into the role of the thumb-like loop in GH-11 xylanases. Protein Eng. Des. Sel. 20:15-23. [DOI] [PubMed] [Google Scholar]
- 30.Payan, F., P. Leone, S. Porciero, C. Furniss, T. Tahir, G. Williamson, A. Durand, P. Manzanares, H. J. Gilbert, N. Juge, and A. Roussel. 2004. The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J. Biol. Chem. 279:36029-36037. [DOI] [PubMed] [Google Scholar]
- 31.Raedschelders, G., C. Debefve, H. Goesaert, J. A. Delcour, G. Volckaert, and S. Van Campenhout. 2004. Molecular identification and chromosomal localization of genes encoding Triticum aestivum xylanase inhibitor I-like proteins in cereals. Theor. Appl. Genet. 109:112-121. [DOI] [PubMed] [Google Scholar]
- 32.Rouau, X., S. Daviet, T. Tahir, B. Cherel, and L. Saulnier. 2006. Effect of the proteinaceous wheat xylanase inhibitor XIP-I on the performances of an Aspergillus niger xylanase in breadmaking. J. Sci. Food Agric. 86:1604-1609. [Google Scholar]
- 33.Saha, B. C. 2003. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30:279-291. [DOI] [PubMed] [Google Scholar]
- 34.Sansen, S., C. J. De Ranter, K. Gebruers, K. Brijs, C. M. Courtin, J. A. Delcour, and A. Rabijns. 2004. Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum xylanase inhibitor-I. J. Biol. Chem. 279:36022-36028. [DOI] [PubMed] [Google Scholar]
- 35.Suen, D. F., and A. H. C. Huang. 2007. Maize pollen coat xylanase facilitates pollen tube penetration into silk during sexual reproduction. J. Biol. Chem. 282:625-636. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, M., A. Kato, N. Nagata, and Y. Komeda. 2002. A xylanase, AtXyn1, is predominantly expressed in vascular bundles, and four putative xylanase genes were Identified in the Arabidopsis thaliana genome. Plant Cell Physiol. 43:759-767. [DOI] [PubMed] [Google Scholar]
- 37.Tahir, T. A., J. G. Berrin, R. Flatman, A. Roussel, P. Roepstorff, G. Williamson, and N. Juge. 2002. Specific characterization of substrate and inhibitor binding sites of a glycosyl hydrolase family 11 xylanase from Aspergillus niger. J. Biol. Chem. 277:44035-44043. [DOI] [PubMed] [Google Scholar]
- 38.Tahir, T. A., A. Durand, K. Gebruers, A. Roussel, G. Williamson, and N. Juge. 2004. Functional importance of Asp37 from a family 11 xylanase in the binding to two proteinaceous xylanase inhibitors from wheat. FEMS Microbiol. Lett. 239:9-15. [DOI] [PubMed] [Google Scholar]
- 39.Törrönen, A., and J. Rouvinen. 1997. Structural and functional properties of low molecular weight endo-1,4-beta-xylanases. J. Biotechnol. 57:137-149. [DOI] [PubMed] [Google Scholar]
- 40.Trogh, I., J. F. Sørensen, C. M. Courtin, and J. A. Delcour. 2004. Impact of inhibition sensitivity on endoxylanase functionality in wheat flour breadmaking. J. Agric. Food Chem. 52:4296-4302. [DOI] [PubMed] [Google Scholar]
- 41.Van Campenhout, S., and G. Volckaert. 2005. Differential expression of endo-β-1,4-xylanase isoenzymes X-I and X-II at various stages throughout barley development. Plant Sci. 169:512-522. [Google Scholar]