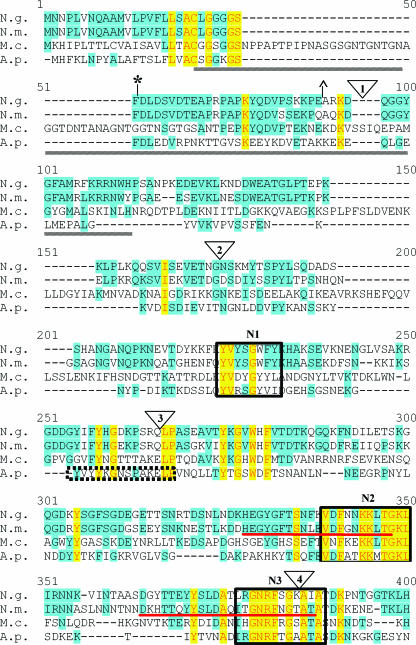
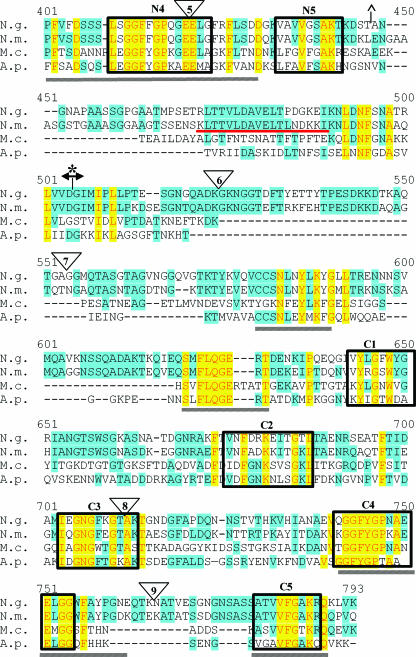
FIG. 1.
Alignment of TbpB proteins from bacterial pathogens. TbpB from N. gonorrhoeae FA19 (N.g.), N. meningitidis M982 (N.m.), M. catarrhalis 4223 (M.c.), and A. pleuropneumoniae serotype 7 (A.p.) were aligned with the program Vector NTI. Identical residues are shaded in yellow while similar residues are shaded in blue. HA epitope fusion points are shown as triangles numbered 1 to 9. Previously identified regions of conservation among TbpB proteins are underlined by gray lines (11). The previously identified denaturation-resistant Tf-binding domains in the TbpB proteins of N. gonorrhoeae (*) and N. meningitidis (^) are flanked by their respective symbols. The double arrowhead indicates the division between the N- and C-terminal halves of the protein. Regions of internal similarity between the N- and C-terminal halves of TbpB are boxed and labeled N1 through N5 and C1 through C5, respectively. The sequence motifs previously identified as important for high-affinity Tf binding of N. meningitidis (35) are underlined in red. The A. pleuropneumoniae peptide shown to bind Tf (43) is denoted by the dotted-line box.