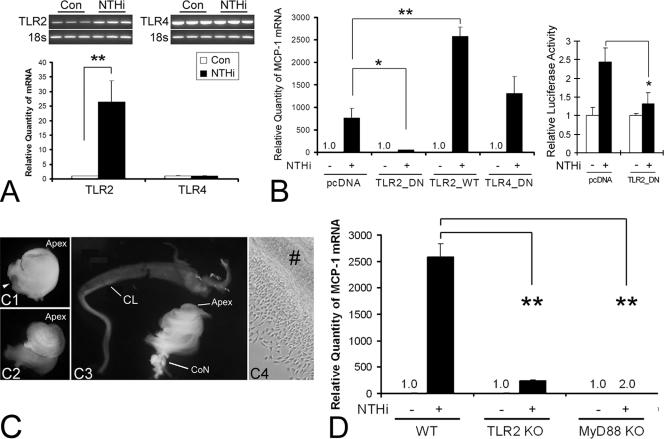
FIG. 3.
TLR2 and MyD88 are involved in NTHI-induced MCP-1 up-regulation of SLFs. (A) Both conventional RT-PCR and real-time quantitative PCR show that the spiral ligament cell line expresses TLR2 and TLR4. TLR2 is up-regulated only by NTHI treatment in SLFs. Con, control. (B) NTHI-induced MCP-1 up-regulation is inhibited by the dominant-negative TLR2 construct (TLR2_DN) but not by the TLR4 dominant-negative construct (TLR4_DN) (left panel). In contrast, overexpression of wild-type TLR2 (TLR2_WT) enhances NTHI-induced MCP-1 up-regulation. The luciferase assay demonstrates that the TLR2 dominant-negative construct inhibits NTHI-induced activation of the MCP-1 promoter (right panel). MCP1-E vector was cotransfected with the TLR2 dominant-negative construct and pRL-TK vector. pcDNA, mock transfection with a blank vector. (C) Dissection microscopic views demonstrating separation of the cochlear lateral wall from the cochlea of gene-targeted mouse pups. The cochlea is isolated with preservation of its normal structure, such as the round window (arrowhead), after dissecting the inner ear from the skull base (C1). After removal of the bony otic capsule (C2), the cochlear lateral wall is separated (C3) and ready for the explant culture (C4). Apex, cochlear apex; CL, cochlear lateral wall; CoN, cochlear nerve; #, an explant of the cochlear lateral wall. (D) Real-time quantitative PCR shows that targeting TLR2 or MyD88 blocks NTHI-induced MCP-1 up-regulation more than 90% and 100%, respectively. The NTHI lysate was incubated with the primary SLFs derived from wild-type or knockout mice after starvation. Cells were harvested 3 h after treatment, and total RNA was extracted. After synthesizing cDNA, multiplex PCR was performed for real-time quantitation, and the CT values of MCP-1 were normalized to the internal control, GAPDH. Results are expressed as fold induction of mRNA quantity or luciferase activity, taking the value of the nontreated group as 1. The experiments were performed in triplicate and repeated more than twice. Values are given as the means ± standard deviations (n = 3). *, P < 0.05; **, P < 0.01.