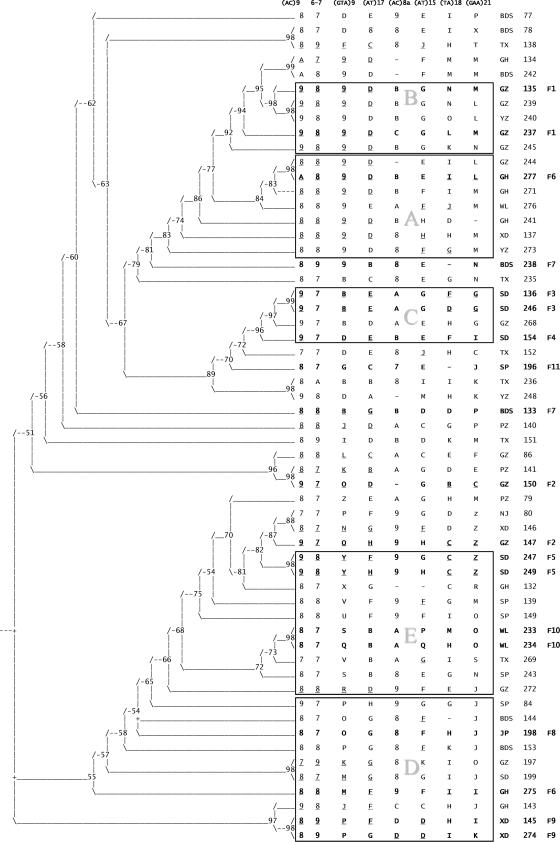
FIG. 2.
Phylogenetic analysis of M. leprae strains isolated from leprosy patients in Qiubei County, Yunnan Province, People's Republic of China. A 50% consensus tree is illustrated, which was obtained with VNTR alleles for eight loci for 57 isolates using parsimony principles as described in Materials and Methods. The numbers at the nodes of branches represent percentages of trees in which the nodes were found. To the right of the tree, the eight locus alleles (repeat number) for each patient are indicated. Alleles 1 to 9 are indicated by the number, while a letter code is used for alleles greater than 9. Alleles 22, 26, 31, 32, 33, 38, 39, 40, 41, and 42 were not found in the data set. Thus, the alleles could be accommodated using 26 letter codes as follows: A to L, 10 to 21, respectively; M to O, 23 to 25, respectively; P to S, 27 to 30, respectively; T to W, 34 to 37, respectively; X and Y, 43 and 44, respectively; and Z, all alleles ≥45. The township of residence is listed after the alleles using capital letters (as described in the legend of Fig. 1), followed by the patient number. The last column indicates the family number for multicase families. The rectangles marked with the letters A to C represent VNTR profiles grouped on the basis of similarity between the strains. The (GTA)9 allele is 9 in clusters A and B and 11 to 13 in cluster C. Strains grouped in clusters D and E are more variable but distinguished by larger (GTA)9 alleles: 15 to 25 in cluster D and >26 in cluster E.