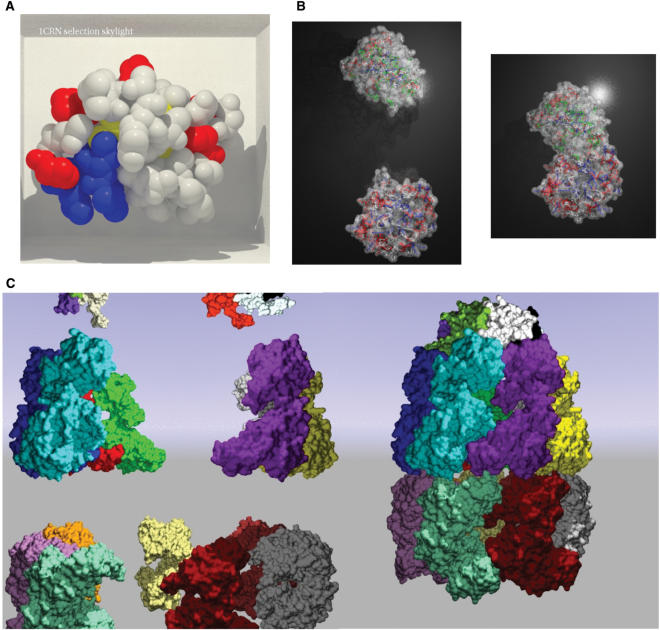
Figure 2.
Example of PMG output. (A) Photorealism: Crambin (PDB code 1CRN) displayed in a box using atom spheres. The rendering is done using the ‘Skylight’ option. (B) Simultaneous multiple representations: two chains isomerase (PDB code 1TIM). The chain A (bottom) is displayed using a backbone spline colored according to the secondary structure, and using a 50% transparent gray surface. A selection of a subset of amino-acids (ASP, GLU, ASN, GLN, LYS and HIS) is drawn using a different representation (ball and sticks colored according to atom charge). The chain B is displayed using a backbone trace colored according to the secondary structure and using a 50% transparent gray surface. A second user selection over different amino-acids (ALA, CYS, PHE, GLY, ISO, LEU and MET) is displayed as ball and sticks and colored according to hydrophobicity. The background is a z-plane textured with material reflection. (C) Large protein assembly animation: This example depicts the oligomerization of the chaperonin Groel–Groes complex, PDB code 1AON (starting position = left image, final position = right image). PMG can handle this large protein assembly (more than 8.000 residues for 21 chains), first by using an ultra wide-angle camera (‘ultra-wide’ option), and then by grouping chains as objects, i.e. by assigning several chains to the same cell, for instance in cell 3.3 Chains: K, L, M and in cell 2.3 Chains: D, E, F. The background is a blue to green sky sphere (‘spheresky’). The complete animation is available at the PMG gallery (http://bioserv.rpbs.jussieu.fr/~autin/help/gallery.html).