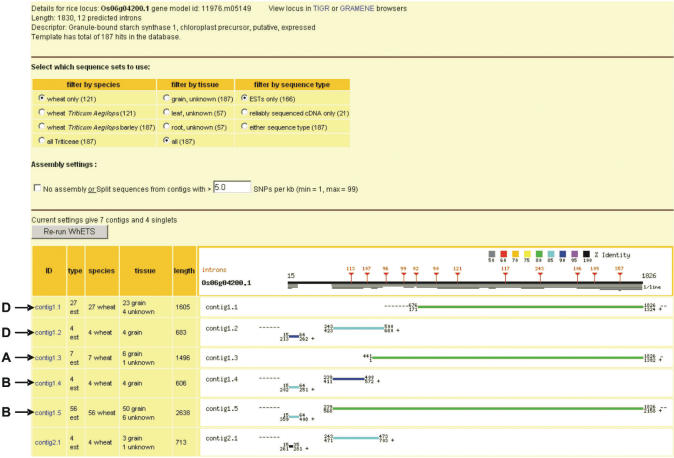
Figure 2.
Output from WhETS for Os06g04200.1. The black line at the top corresponds to the rice gene CDS with intron position and size indicated as red vertical lines with triangles. Thin horizontal lines below this indicate the coverage of hits from the Triticeae sequences. The rows below show these hits, with blast HSPs for contigs and singlets shown as lines coloured according to percentage identity, and coordinates aligned to the rice template. The CAP3 step gives three contigs; one of these (contig 1) is then divided into five new contigs by the SNP analysis step (contigs 1.1, 1.2, etc.) The genome of origin (A, B, D) has been added to the screenshot according to 100% identity matches of the contig consensus to the known-homoeologue transcript sequences (exons of accessions AB019622, AB019623 and AB019624). Contigs 1.1 and 1.2 are not combined because of a lack of substantial overlap. Contigs 1.4 and 1.5 appear to be splice variants with an indel in the 5′UTR. Contigs 2 and 3 appear quite different and may be paralogues.