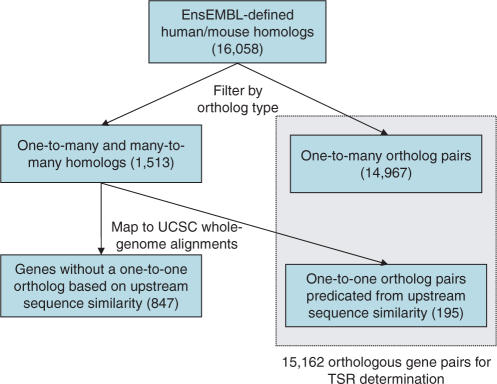
Figure 1.
Determination of one-to-one orthologs for human and mouse genes. An initial set of homologs was downloaded from EnsEMBL v41 (30). All homologs annotated as ‘one2one’ are extracted. To select the closest putative ortholog pairs from homologs with ‘one2many’ or ‘many2many’ relationships, we check for upstream conservation using the whole-genome human–mouse alignments (6). We re-annotate unambiguously aligned homologs as putative one-to-one orthologs, adding 195 gene pairs to our set and bringing the total number of orthologs to 15 162.