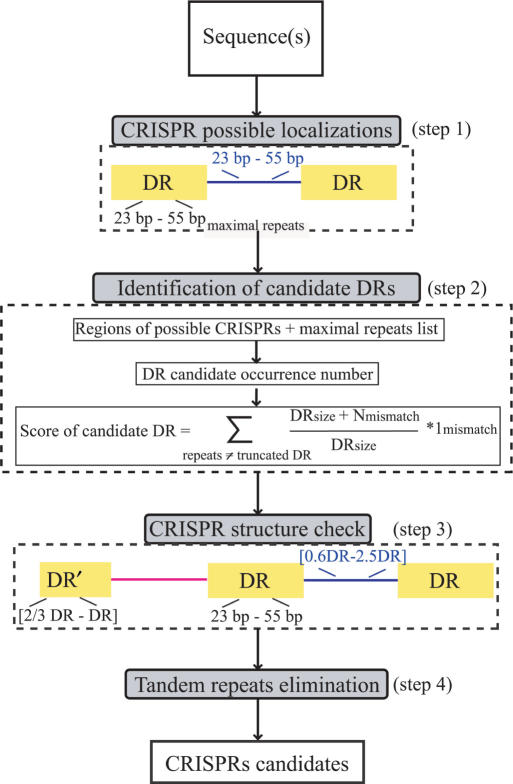
Figure 1.
CRISPR Finder flow chart. (Step 1) Browsing the maximal repeats to get possible CRISPR localizations using the Vmatch program. (Step 2) Consensus DR selection according to candidate occurrences and a score computation: the score privileges internal mismatches between direct repeats of a cluster rather than boundary mismatches. (Step 3) DR and spacers size check. (Step 4) Tandem repeats elimination using ClustalW for aligning spacers.