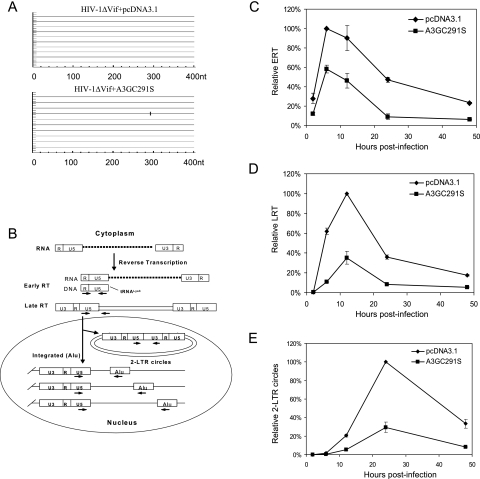
FIG. 1.
Effects of A3GC291S on HIV-1 reverse transcription and nuclear import during a single-round infection of HeLa cells. HIV-1ΔVif was cotransfected with the pcDNA3.1 or A3GC291S expression vector into 293T cells along with the VSV-G expression vector. At 72 h posttransfection, viruses were collected and used to infect HeLa cells. The infected cells were harvested at different time points. (A) A3GC291S is defective for cytidine deamination of HIV-1 DNA. The proviral DNA was extracted at 16 h after infection, and fragments of HIV-1 in the region of the Nef-U5 from virus-infected cultures in the presence of A3GC291S or a control vector were amplified, cloned, and sequenced. G-to-A mutations are shown. (B) Schematic of the early events in HIV-1 replication and the locations of PCR primers. HIV-1 genomic RNAs were reverse transcribed to yield linear viral DNAs. Of these, some were degraded, some were circularized to 1- or 2-LTR circles, and some were integrated. Arrows indicate the approximate locations of the PCR primers used to detect the different HIV-1 DNA forms. Real-time PCR was used for time course analysis of HIV-1 ERT (C), LRT (D), and 2-LTR circle DNA formation (E). nt, nucleotide.