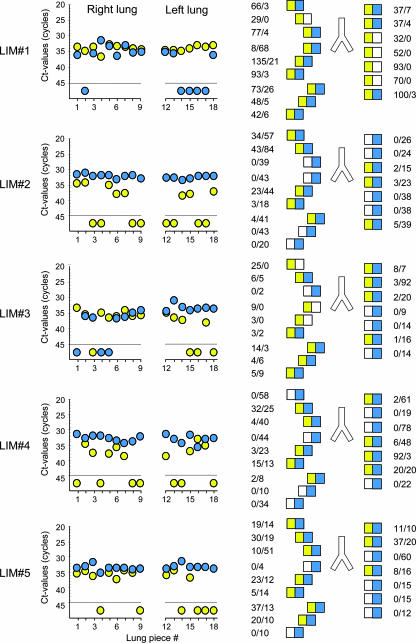
FIG. 3.
Contextual analysis of MIE locus transcription patterns in latently infected lungs. The BALB/c mouse model of syngeneic bone marrow transplantation and infection with the mCMV wild-type Smith (ATCC VR-194) strain was employed to establish viral latency in the lungs of transplantation recipients (41). Transcription analysis for five latently infected mice (designated LIM#1 through LIM#5) was performed at 12 months after transplantation and at 24 h after the in vivo activation of MIE gene expression by 1 μg of recombinant murine TNF-α administered intravenously (40). For the analysis of variegated MIE gene expression, lungs were cut into pieces, specifically into nine pieces derived from the superior, middle, and inferior lobes of the right lung and seven pieces derived from the left lung. Two pieces (pieces 10 and 11) of the postcaval lobe were used to control for the presence and load of latent viral DNA (not shown). Transcripts IE1 and IE2 were quantified for each of the total number of 80 tissue pieces by the respective real-time RT-PCRs from 10% aliquots of the yields of total RNA purified with a QIAGEN RNeasy Plus kit. (Left panel) raw data given as CT values for the 16 lung pieces (pieces 1 to 9 and 12 to 18) tested per mouse. The dashed line marks the cutoff CT value separating negative from positive samples. Data for IE1 and IE2 are shown as yellow- and blue-filled circles, respectively. (Right panel) corresponding lung pictograms in anatomical view. Results are expressed in numbers of IE1/IE2 transcripts per test aliquot. In accordance with the MIE locus architecture illustrated in Fig. 1, yellow and blue boxes symbolize the presence of IE1 and IE2 transcripts, respectively. Open boxes symbolize the absence of transcripts.