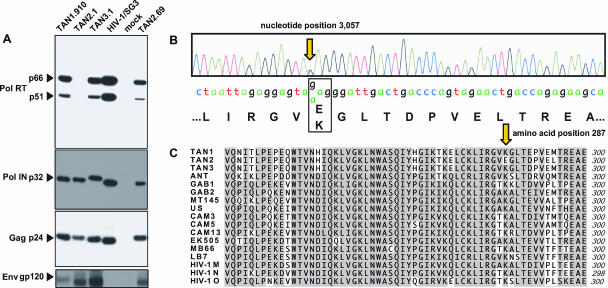
FIG. 6.
Identification of an inactivating mutation in the TAN2.1 pol gene. (A) Western blot analysis of transfection-derived virions using Pol-, Gag-, and Env-specific antibodies. Protein loading was equilibrated by RT activity, with 100 ng loaded to detect Pol and Gag proteins and 400 ng loaded to detect Env (TAN2.1 was used at maximal RT activity). Bands corresponding to RT (p66 and p51), integrase (p32), capsid (p24), and the extracellular envelope (gp120) protein are indicated. Lane 4 lacks a gp120 band because the HIV-1/SG3 Env protein failed to cross-react with the SIVcpzPts-specific gp120 antiserum. Minor p32 migration differences correspond to predicted molecular weight differences of integrase proteins from the different viruses. (B) Sequence chromatogram of the TAN2 pol gene revealing a site of sequence ambiguity (G/A double peak) at position 3057 with differential coding potential (E or K at amino acid position 287 of the RT protein). (C) Alignment of available SIVcpzPts (TAN1, TAN2, TAN3, and ANT), SIVcpzPtt (GAB1, GAB2, MT145, US, CAM3, CAM5, CAM13, EK505, MB66, and LB7), and HIV-1 (group consensus) RT protein sequences (position 241 to 300) available from the Los Alamos HIV Sequence Database. Amino acids identical among all RT sequences are shaded, with position 287 highlighted by an arrow.