Figure 1.
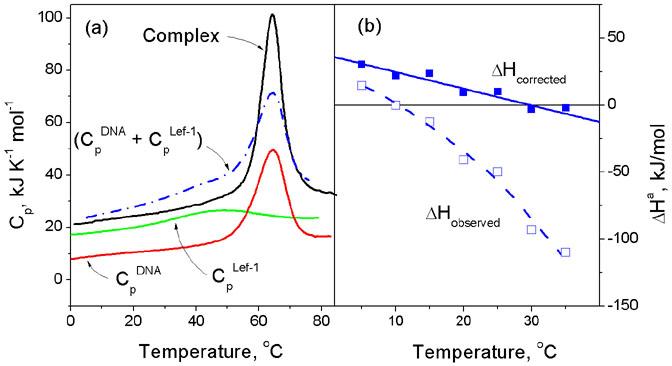
(a) The heat capacity functions of a typical DBD, the HMG box from LEF-1, its target DNA duplex and their complex, determined by differential scanning calorimetry (DSC). This shows that the protein starts to unfold from very low temperatures but on association with DNA it refolds and forms a stable complex that dissociates and unfolds cooperatively at about 62°C. The heat of protein refolding at any given temperature can be determined by integrating the differences between the summed heat capacity of the free protein and DNA (Blue dot-dash line) and the observed heat capacity of the complex (Black solid line). (b) The observed enthalpy of association of the HMG box from LEF-1 with its target DNA measured by isothermal titration calorimetry (ITC) and the function corrected for heats of protein refolding upon binding: the corrected function corresponds to the enthalpy of association of the fully folded DBD with the DNA. For details see4,7.
