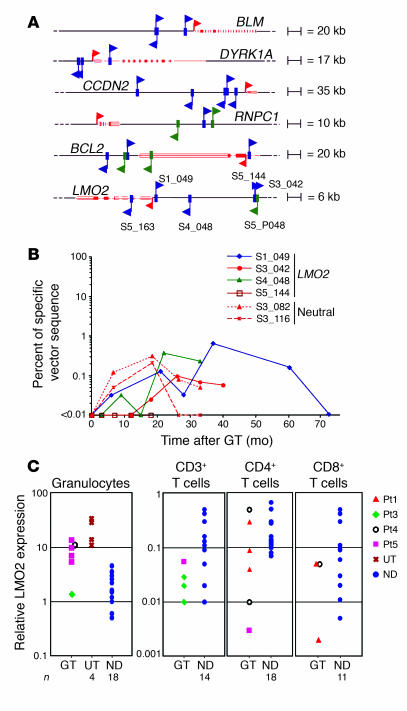
Figure 2. RIS hot spots in patients with ADA-SCID without clonal expansion or overexpression.
(A) Schematic representation of RISs in the most frequently hit genes. Additional RISs outside these intervals in the proximity of CCND2 and BCL2 are also indicated. In vitro RISs are shown as green arrows and in vivo RISs as blue arrows; the direction of the arrow denotes orientation of the retroviral vector. The upper or lower position of the arrow refers to an LTR-genomic junction sequence mapping to the positive (above line) or negative (below line) chromosomal strand, respectively. Genes are depicted in red, with filled boxes and red arrows indicating exons and TSSs, respectively. RISs in the LMO2 locus were detected in T cells (Pt1, S1_049, 27 months after GT; Pt3, S3_042, 18 months after GT; Pt4, S4_048, 26 months after GT), granulocytes (Pt5, S5_144 and S5_163, 6 and 12 months after GT, respectively), and in Pt5 preinfusion CD34+ cells (S5_P048). (B) Quantification of the LMO2-containing clones within the total T cell population. The frequency of insertions was measured by sequence-specific real-time PCR in purified CD3+ T cells isolated at different time points after GT. Dotted lines show the frequencies of nonrecurrent (neutral) RISs retrieved from Pt3 as a control of physiologic clonal fluctuations. (C) Expression of LMO2 in peripheral blood T cells and granulocytes. Total RNA was isolated from the indicated cell subsets of GT-treated patients, patients with ADA-SCID undergoing polyethylene glycol-modified bovine ADA treatment (UT, not treated with GT), or age-matched healthy controls (ND). LMO2 gene expression was measured by quantitative RT-PCR using the standard curve of a reference sample and normalized for the expression of the housekeeping gene HPRT.