Figure 2.
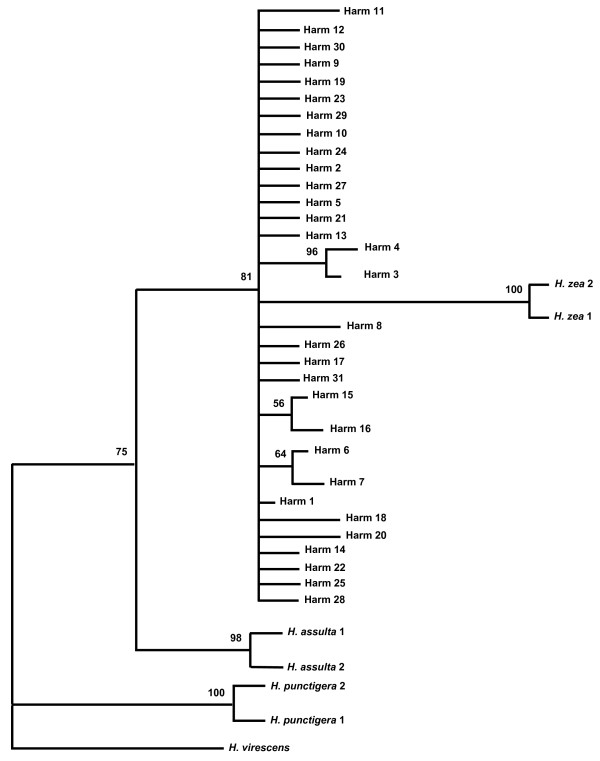
Maximum Likelihood (ML) tree of H. armigera (Harm-1 to Harm-31), H. zea (Hzea-1, Hzea-2), H. assulta and H. punctigera based on partial COI haplotypes sequences. Numbers above the nodes indicate bootstrap support. The outgroup used was Heliothis virescens. The inclusion of additional haplotypes Harm-32, Harm-33, and Hzea-3 to Hzea-11 did not alter the overall topology, and bootstrap values of the ML tree after 1,000 bootstrap replications remained high, with all H. zea haplotypes confidently clustered (bootstrap value = 96) within the H. armigera clade. H. punctigera remained basal to H. assulta (bootstrap value = 99), and the H. armigera/H. zea clade (bootstrap value = 78) shared a most common ancestor with H. assulta (bootstrap value = 97) (data not shown).
