Abstract
Epithelia in animals are colonized by complex communities of microbes. Although a topic of long-standing interest, understanding the evolution of the microbial communities and their role in triggering innate immune responses has resisted analysis. Cnidaria are among the simplest animals at the tissue grade of organization. To obtain a better understanding of the microbiota associated with phylogenetically ancient epithelia, we have identified the epibiotic and endosymbiotic bacteria of two species of the cnidarian Hydra on the basis of rRNA comparisons. We analyzed individuals of Hydra oligactis and Hydra vulgaris from both laboratory cultures and the wild. We discovered that individuals from both species differ greatly in their bacterial microbiota. Although H. vulgaris polyps have a quite diverse microbiota, H. oligactis appears to be associated with only a limited number of microbes; some of them were found, unexpectedly, to be endosymbionts. Surprisingly, the microfauna showed similar characteristics in individuals of cultures maintained in the laboratory for >30 years and polyps directly isolated from the wild. The significant differences in the microbial communities between the two species and the maintenance of specific microbial communities over long periods of time strongly indicate distinct selective pressures imposed on and within the epithelium. Our analysis suggests that the Hydra epithelium actively selects and shapes its microbial community.
Keywords: endosymbiosis, epithelial defense, innate immunity, microbial community, host–microbe interaction
Epithelia of all animals are colonized by complex communities of microbes (1). Different epithelia within one organism have different microbial communities (2). Perturbations and imbalances in this usually beneficial relationship underlie many human diseases. In the absence of bacterial colonization, germ-free animals display defects in the ability to fight infections by pathogenic bacteria and viruses (3). The mechanisms, which mediate the interdependent and complex interactions within microbial communities and the host epithelium as well as the influence of the microbiota on immune functions, are not yet discovered. Molecular analysis of the bacterial microbiota in the human stomach (2) recently uncovered an unprecedented bacterial diversity that is different from the bacteria found in mouth or esophagus. In a reciprocal transplantation experiment with the microbial community of mice and zebrafish, evidence was provided (4) that the gut epithelium is actively shaping the microbiota. To recognize and manage these complex communities of microbes, vertebrates, in contrast to invertebrates, are thought (5) to have evolved adaptive immunity.
The microbiota living on or in invertebrate epithelia and their influence on the host immune system are largely unknown. Are there identifiable core microbiota associated with a given host species? How are the microbiota selected, and how did they evolve within and between hosts? Here we characterize bacterial diversity within the epithelia of the cnidarian Hydra. Cnidaria are one of the earliest branches in the animal tree of life; they represent the simplest animals at the tissue grade of organization (Fig. 1A) and have a body plan in which there is no physical barrier between the host tissue and the microbes. Thus, Cnidaria occupy an important evolutionary position for understanding direct host–microbe interactions. We have shown recently that the cnidarian immune system has evolved an elaborate mechanism to delete or suppress pathogenic intruders (6). Research into molecules of the innate immune response in Cnidaria (6) has identified pattern-recognition receptors, most notably the Toll-like receptors, allowing permanent surveillance of resident microbiota and intruding pathogens. Moreover, two closely related species of Hydra, Hydra oligactis (Fig. 1B) and Hydra vulgaris (Fig. 1C), are remarkably different in their stress response (7, 8). These differences appear to be correlated with differences in the antimicrobial activity against different bacteria (T.C.G.B., unpublished data). Because this correlation appears to be caused by differences in selective constraints, we asked whether the species-specific differences in the immune and stress response are reflected by the microbiota associated with these two Hydra species and whether there is an identifiable specific core microbiota associated with a given species.
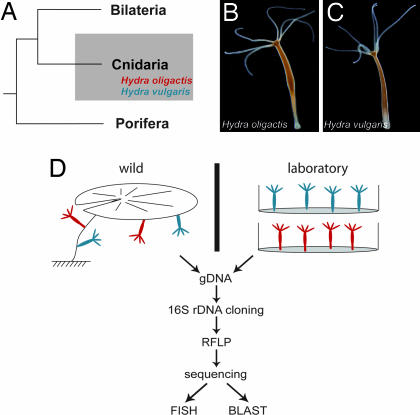
Fig. 1.
Analysis of Hydra-associated bacteria. (A) Phylogenetic position of the cnidarian Hydra. (B and C) Morphological characteristics of the two Hydra species analyzed. (D) Schematic representation of the approach. Bacterial microbiota were compared between H. vulgaris (blue) and H. oligactis (red) from laboratory culture (Right; drawn in plastic dishes) and the wild (Left; attached to water lily).
Results
Microbiota in Two Closely Related Hydra Species.
To identify the microbiota in the Hydra epithelium, we performed a comprehensive bacterial DNA analysis in individuals of two Hydra species, H. oligactis (Fig. 1B) and H. vulgaris (Fig. 1C). Both species are closely related (9) and located within the phylum Cnidaria (Fig. 1A). We compared 10 individuals from each species that were kept for >30 years under identical laboratory culture conditions with 10 H. oligactis individuals directly isolated from Lake Pohlsee, 6 H. oligactis individuals isolated from Lake Ploen, and 6 H. vulgaris individuals isolated from Lake Pohlsee. The experimental procedure is shown in Fig. 1D. For bacterial genotyping, 16S rRNA genes were amplified by PCR from each sample and cloned. From each sample, ≈46 clones (see Table 1) were randomly selected for restriction fragment length polymorphism (RFLP). A total of 68 16S rRNA sequences formed the final analyzed data set.
Table 1.
Bacterial species richness in Hydra
| n | S | Chao1 |
||
|---|---|---|---|---|
| Mean ± SD | 95% CI | |||
| Ho (lab) | 77 | 3 | 4 ± 2 | (3–16) (a) |
| Ho (Lake Pohlsee) | 34 | 4 | 5 ± 2 | (4–17) (a, b) |
| Ho (Lake Ploen) | 54 | 5 | 5 ± 0 | (–5) (a) |
| Hv (lab) | 44 | 12 | 17 ± 6 | (13–41) (b, c) |
| Hv (Lake Pohlsee) | 39 | 12 | 19 ± 8 | (13–54) (c) |
| Ho (medium) | 43 | 25 | 43 ± 12 | (30–85) (d) |
| Hv (medium) | 46 | 22 | 35 ± 10 | (26–70) (d) |
| Lake Pohlsee (water) | 43 | 23 | 30 ± 6 | (25–51) (d) |
Hv, H. vulgaris; Ho, H. oligactis; lab, animals taken from laboratory culture; Lake Pohlsee, animals taken from Lake Pohlsee; Lake Ploen, animals taken from Lake Ploen; n, total no. of analyzed clones; S, identified bacterial phylotypes; Chao1, richness estimator. Significant differences are indicated by different letters.
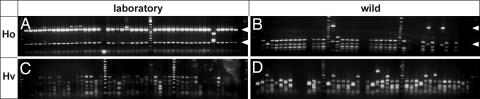
As shown in Fig. 2, the restriction fragment length polymorphism patterns revealed drastic differences between the two Hydra species. In 75 of 77 clones from H. oligactis from the long-term laboratory culture (Fig. 2A), we detected one dominant RFLP pattern, whereas in H. vulgaris individuals from the laboratory culture (Fig. 2C) 16 different RFLP patterns could be observed. The individuals from these cultures were exposed to constant “environmental” conditions, including culture medium, food, and temperature for >30 years. Detecting drastic differences in the bacterial communities was, therefore, completely unexpected and indicated differences in selective constraints. To determine whether similar differences can be observed in Hydra individuals directly isolated from the wild, we performed bacterial genotyping in H. oligactis and H. vulgaris individuals isolated from two different lakes near Kiel. As also shown in Fig. 2, the RFLP patterns from 6 individuals of H. vulgaris isolated from Lake Pohlsee (Fig. 2D) show complexity similar to the RFLP pattern in the laboratory animals (Fig. 2C). Moreover, and strikingly similar to the laboratory animals, in the H. oligactis polyps isolated from the lake (Fig. 2B), only one dominant RFLP pattern could be observed. Interestingly, although the bacterial RFLP pattern in the majority of H. oligactis polyps (72 of 89 clones analyzed) from the wild seems to be different from the dominant RFLP pattern in the corresponding laboratory culture (see arrows in Fig. 2 A and B), seven bacterial RFLP patterns from H. oligactis polyps from the wild show an RFLP pattern identical to those from H. oligactis polyps from the long-term laboratory cultures. Our results demonstrate not only that H. vulgaris polyps have a quite diverse microbial fauna whereas H. oligactis appear to be associated with only a limited number of microbes, but also that intraspecies differences in the RFLP patterns of the samples from the wild and the long-term laboratory culture are much smaller than the differences between the two species.
Fig. 2.
RFLP analysis of 16S rRNA genes of bacteria associated with two different Hydra species. (A) H. oligactis from the laboratory culture. (B) H. oligactis isolated from the wild (Lake Pohlsee). (C) H. vulgaris from the laboratory culture. (D) H. vulgaris isolated from the wild (Lake Pohlsee). Ho, H. oligactis; Hv, H. vulgaris.
Phylogenetic Analysis Reveals Species-Specific Phylotypes.
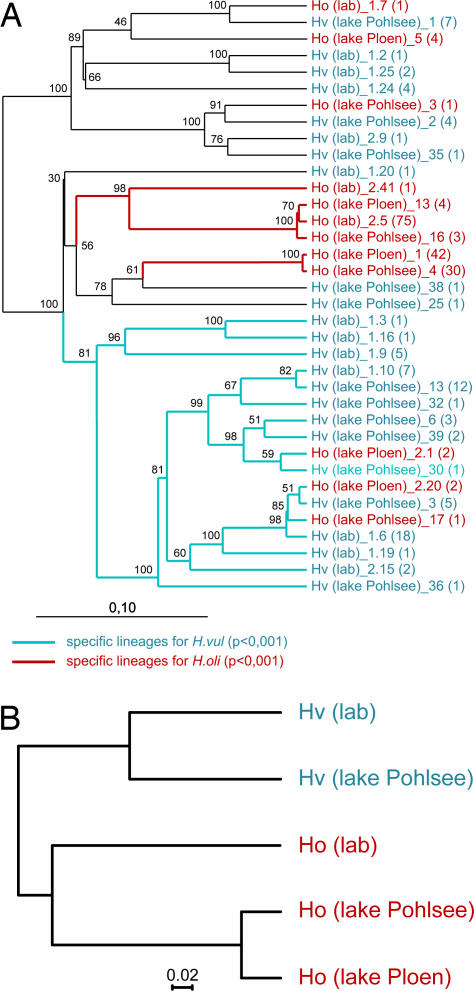
To identify the bacterial divisions in the H. oligactis and H. vulgaris epithelium, partial sequences (≈1,450 bp) were obtained from each RFLP type. After phylogenetic analyses were performed, sequences with ≥97% similarity were designated phylotypes. Fig. 3A shows the phylogenetic tree with the 36 phylotypes identified in the sequence data set. In the H. vulgaris samples from the laboratory and the wild, we identified in each case 12 different bacterial phylotypes. In H. oligactis, we identified 3 phylotypes in the laboratory culture and 4 and 5 phylotypes, respectively, in the samples from the two lakes.
Fig. 3.
Phylogenetic comparison of identified bacterial phylotypes from the different Hydra species. (A) Neighbor-joining tree (Olsen correction) with the 36 identified 16S rDNA phylotypes from five different samples. The number of RFLP patterns within each phylotype is shown in parentheses. Bootstrap values are shown at the corresponding nodes (n = 100). [Scale bar: evolutionary distance (0.1 substitution per nucleotide).] Specific bacterial lineages for the different Hydra species analyzed with UniFrac are indicated in blue (specific for H. vulgaris, P < 0.001) and red (specific for H. oligactis, P < 0.001). (B) Jackknife environment cluster tree (weighted UniFrac metric, based on the 36-sequence tree; ref. 10) of the analyzed bacterial communities. One hundred jackknife replicates were calculated, and each node was recovered with >99.9%. (Scale bar: distance between the environments in UniFrac units.) Hv, H. vulgaris; Ho, H. oligactis; lab, animals from laboratory culture; lake Pohlsee, animals taken from Lake Pohlsee; lake Ploen, animals taken from Lake Ploen.
To determine whether the observed microbiota of the two Hydra species are species-specific, we processed the data using two independent computational approaches. First, the bacterial communities of H. vulgaris and H. oligactis were compared using the UniFrac computational tool (10). The UniFrac significance (using 1,000 iterations; P = 0.04) revealed a significant difference between both species. Additionally, the UniFrac lineage-specific analysis, in which the abundance of each phylotype (Fig. 3A, in parentheses) is considered, identified specific bacterial divisions for the two different Hydra species (P < 0.001), which indicates that the composition of the bacterial guilds is different in the two different species. Next, we tested the validity of these conclusions by performing UniFrac hierarchical cluster analysis (also known as UPGMA) on the environments based on a distance matrix that is generated by calculating pairwise UniFrac values. As shown in Fig. 3B, the H. vulgaris data sets from both the laboratory culture and the wild clearly segregate from the data sets obtained from H. oligactis. These facts attest to the resilience of the bacterial communities associated with the different Hydra species and indicate drastic compositional differences.
In the second approach, we used EstimateS (version 8; http://purl.oclc.org/estimates) to gain insight into the bacterial species richness. As shown in Table 1, estimation of the number of phylotypes associated with both Hydra species by the nonparametric Chao1 algorithm (11) also revealed significant differences in the microbiota associated with both species. In the H. vulgaris samples from the laboratory and the wild, we could identify 12 different phylotypes in each case. Therefore, Chao1 estimates a mean phylotype number of 17 and 19 for the H. vulgaris samples from the laboratory and the wild, respectively. The number of identified phylotypes associated with H. oligactis was, in all three samples, significantly smaller than the one in H. vulgaris. As shown in Table 1, in H. oligactis from the laboratory we could identify three different phylotypes, whereas in the H. oligactis samples taken from the lakes, four (Lake Pohlsee) and five (Lake Ploen) phylotypes were detected. Thus, the species richness of bacteria appears to be lower in H. oligactis than in H. vulgaris. Moreover, phylotypes (RFLP patterns) associated with Hydra tissue could not be detected in the surrounding water.
Identification of Multiple Bacteria Species within the Hydra Epithelium.
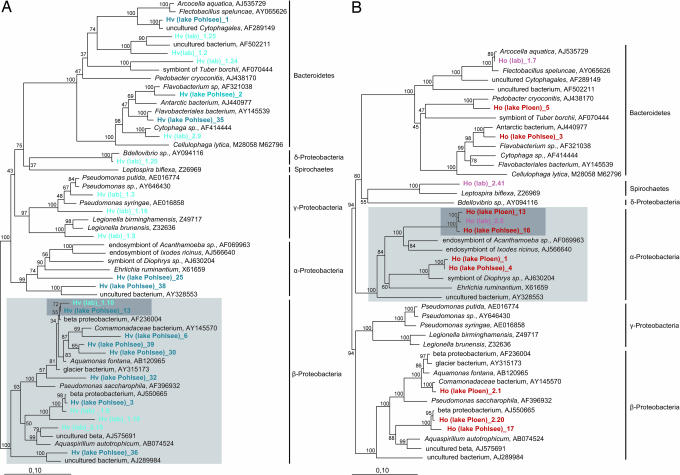
To characterize the bacterial community, we analyzed the microbiota based on nearest relative (as determined by BLASTN search) and phylogenetic affiliation. As shown in Fig. 4, the 36 identified phylotypes represent three different bacterial divisions and are dominated by Proteobacteria and Bacteroidetes. In H. vulgaris (Fig. 4A), the α-Proteobacteria are completely absent in the epithelium from laboratory-cultured polyps, whereas γ-Proteobacteria (Pseudomonadales) are conspicuously absent in the samples from the wild. The Bacteroidetes and β-Proteobacteria are represented by several phylotypes in H. vulgaris tissue collected from both the laboratory and the wild. Interestingly, within the β-Proteobacteria, one phylotype [Hv(lab)_1.10 / Hv(Lake Pohlsee)_13] (Fig. 4A, gray shadowed) appears to be species-specifically associated with H. vulgaris. Strikingly, this H. vulgaris-specific bacterial phylotype was detected not only in the polyps from the long-term laboratory culture but also in H. vulgaris polyps directly isolated from the wild, which indicates that H. vulgaris actively maintains the association with this bacterial species. Moreover, and in sharp contrast to the data set obtained from H. oligactis (Fig. 4B), members of the β-Proteobacteria appear to be abundant in H. vulgaris tissue.
Fig. 4.
Phylogenetic position (16S rRNA gene sequences, neighbor-joining tree) of identified phylotypes. (A) Phylogenetic position of identified bacterial phylotypes associated with H. vulgaris. (B) Phylogenetic position of identified bacterial phylotypes associated with H. oligactis. Light gray shadowed bacterial groups indicate species-specific bacterial guilds; dark gray shadowed bacterial phylotypes indicate species-specific bacterial species. The branch length indicator displays 0.1 substitution per site.
In the bacterial phylotypes associated with H. oligactis (Fig. 4B), the majority of phylotypes belong to the α-Proteobacteria (Rickettsiales). The molecular analysis shown in Fig. 4B also indicates that within the α-Proteobacteria, one phylotype appears to be species-specific for H. oligactis. This bacterial species was identified both in the sample from the laboratory culture and in polyps isolated from the wild. In H. oligactis, no phylotypes could be identified belonging to the γ-Proteobacteria.
Taken together, these data show that both Hydra species select particular bacterial guilds. H. oligactis and H. vulgaris maintain these species-specific bacterial communities even when cultured under constant conditions for >30 years. Alternatively, it seems possible that the bacteria are also actively involved in selecting the host.
Discovery of Endosymbiotic Bacteria in Epithelial Cells of H. oligactis That Are Absent in H. vulgaris.
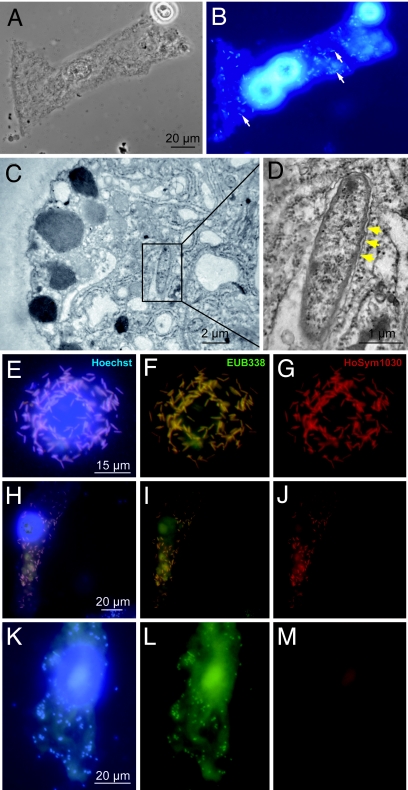
Unexpectedly, the sequence analyses shown in Fig. 4B indicated that the H. oligactis-specific bacterial phylotype was most closely related to endosymbiotic bacteria. Because it was not known so far whether H. oligactis polyps contain endosymbionts and, if so, in which cells they were located, we analyzed the epithelial cells for the presence of endosymbiotic bacteria. The microscopic analysis (Fig. 5 A and B) revealed numerous bacteria within all epithelial cells in all H. oligactis polyps analyzed irrespective of whether the animals were taken from the long-term laboratory culture (n = 15) or directly from the wild (n = 5). In contrast, no bacteria could be detected within the epithelial cells of H. vulgaris (data not shown). Transmission electron microscopy (Fig. 5 C and D) revealed rod-shaped bacteria of 2–4.5 μm within the cytoplasm of H. oligactis ectodermal epithelial cells. As shown in Fig. 5D, the endosymbiotic bacteria are surrounded by a secondary membrane and are similar in morphology to bacteria identified previously as symbiotic bacteria in Acanthamoeba (12) or Ixodes (13).
Fig. 5.
Microscopic analysis of endosymbiotic bacteria in H. oligactis. Macerated epithelial cell was stained with Hoechst and evaluated with phase-contrast microscopy (A) and epifluorescence microscopy (B). (C and D) Transmission electron micrographs of bacterial endosymbionts in the cytoplasm of ectodermal epithelial cell. Secondary membrane is indicated by yellow arrowheads. (E–M) In situ hybridization (FISH) reveals endosymbiont identity. (E–G) Trypsin-digested epithelial cells. (H–M) Macerated epithelial cells. (E, H, and K) Cells were stained with Hoechst. (F, I, and L) Bacteria cells were stained with the fluorescently labeled oligonucleotide probe EUB338. (G, J, and M) Bacterial cells were stained with the phylotype-specific probe HoSym1030. All cells were viewed with epifluorescence microscopy and appropriate filter sets.
To define the identity of the bacterial endosymbiont in H. oligactis, epithelial cells from animals cultured under laboratory conditions were subjected to whole-cell hybridization with fluorescence-labeled oligonucleotide probes. As shown in Fig. 5, all Hoechst-stained bacteria (Fig. 5E) hybridized with a Eubacteria-specific probe (EUB338) (Fig. 5F). Fig. 5G indicates that all of these bacteria hybridize also with a probe (HoSym1030), which is specific for the H. oligactis-specific bacterial phylotype described above (see Fig. 4B).
Next, we examined whether this putative symbiont was also present in epithelial cells of H. oligactis from the wild. Most strikingly (and shown in Fig. 5J), bacteria in ectodermal epithelial cells of H. oligactis isolated directly from the wild also hybridized to probe HoSym1030, indicating that the bacterial symbionts in polyps from the laboratory and the wild are closely related. However, in contrast to H. oligactis polyps from the long-term laboratory culture (Fig. 5 E–G), in polyps isolated from the wild only a subpopulation of epithelial cells (≈20%) were found to harbor the putative symbiont detected by the HoSym1030 probe (Fig. 5 H–J). As also indicated in Fig. 5L, all epithelial cells in H. oligactis polyps from the wild did contain endosymbiotic bacteria because they were detected by the eubacterial-specific probe EUB338. Thus, both H. oligactis from the long-term laboratory culture and from the wild do contain endosymbiotic bacteria. Polyps from the wild appear to contain at least two different endosymbiotic bacteria species (one of them is identical to the symbiont in the laboratory culture), whereas epithelial cells from the laboratory culture appear to contain a homogeneous population of a single endosymbiotic bacteria species.
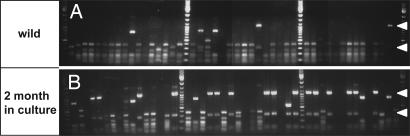
Transition from Lake to Laboratory Causes a Shift in the Bacterial Community in H. oligactis.
To determine the impact of different environmental conditions on the bacterial community in H. oligactis, we cultured some of the polyps that were taken from the wild for 2 months under standard laboratory conditions and thereafter compared the bacteria-specific 16S rDNA RFLP pattern. As shown in Fig. 6, culturing of H. oligactis from the wild under laboratory conditions has drastic effects on the composition of the bacterial community. Although species of the α-Protobacteria that are the dominant species in the long-term culture (indicated by arrowheads in Fig. 6) can also be found in polyps 2 months and 6 months (data not shown) after the shift to the laboratory, other bacteria disappear from the tissue collected in the wild. Fig. 6B also indicates the presence of microbes in the samples from the short-term cultures that were not detected in the samples from the wild. Thus, H. oligactis not only is associated with species-specific symbiotic eubacteria but also responds to changes in the environment with changes in the bacterial community. Even under long-term and constant culture conditions, however, this bacterial community is very different from the community detected in H. vulgaris (see also Fig. 2B). Thus, Hydra appear to select their specific bacterial microbiota.
Fig. 6.
Comparative RFLP analysis of bacterial 16S rRNA genes associated with H. oligactis isolated from Lake Ploen (A) and after cultivation in the laboratory for 2 months (B).
Discussion
Because all epithelia in all animals, including man, appear to be colonized by microbial communities, it becomes important to understand the general principles by which these microbial communities evolve. In this work, we describe the bacterial microbiota in epithelia from animals belonging to one of the earliest branches in the animal tree of life. Importantly, we show that in these animals the microbial communities are species-specific. The unexpected observation that microbial communities were similar in Hydra polyps taken from the wild and polyps cultured for >30 years in the laboratory points to coadaptive radiation of Hydra and its microbial communities.
In the absence of migratory phagocytic cells and any physical barrier between the epithelium and the microbes, the epithelium of the freshwater polyp Hydra is remarkably well equipped to survive in an environment teeming with potential pathogens (6). The current work used molecular tools to reveal the previously uncharacterized bacterial microbiota in two species of Hydra. The Hydra epithelium was found to be colonized by a complex and dynamic community of microbes, and individuals from both species differed greatly in their microfauna. We also found that the bacterial species richness is significantly larger in H. vulgaris than in H. oligactis. Although our data show that the composition of the specific microbial communities associated with H. oligactis may vary in time (Fig. 6), we observed an unexpected degree of similarity between the bacterial composition in individuals of laboratory cultures and polyps directly isolated from the wild (Fig. 3B). This observation indicates that the identified bacteria reflect resident species rather than transient “tourists” passing through with food, water, and other environmental components. The differences in the microbial communities between the two species and the maintenance of specific microbial communities over long periods of time strongly indicate distinct selective pressures imposed on and within the Hydra epithelium and suggest that the epithelium actively selects and shapes its microbial community.
Although this pilot study is far from being quantitative, it indicates that the number of bacterial species permanently associated with epithelia in Hydra is much less than, e.g., the number of resident bacterial species found in the human stomach or mouth (2). This finding reinforces the idea (5) that the diversity of resident bacterial communities is more complex in vertebrates than in invertebrates and that this difference may have prompted the evolution of a memory-based adaptive immune system in vertebrates.
Because in Hydra (T.C.G.B., unpublished data) as in vertebrates (14), differences in microbiota are correlated with differences in immune functions, it appears that epithelia may have an evolutionary requirement for the specific immunomodulatory direction provided by the microbiota. The observation (15) that in the absence of bacteria Hydra polyps show strong developmental defects and are unable to proliferate asexually by budding and that normal budding is resumed with nonsterile food or medium reinoculated with bacteria, supports the view that bacterial microbiota play a pivotal role in Hydra. It now will be important to assess the impact of the different microbiota on the innate immune system and to identify the environmental factors operating to select a specific bacterial community.
Depletion of microbiota appears to have profound effects in health and well-being in both Hydra and humans. Thus, in both Hydra and humans, the immune system is likely to be strongly affected by the need to maintain a substantial resident microbiota. Genetic defects in the ability of the human immune system to respond adequately to the epithelial microbiota appear to predispose individuals to inflammatory diseases (14). The results shown above provide compelling evidence for a complex cross-talk of an epithelial barrier and the residing microbiota at a basal level of evolution. In Hydra, this cross-talk is complemented by unique biological and experimental opportunities. Thus, studies in Hydra may provide a paradigm for the characterization and analysis of microbial communities and their important, as-yet undiscovered roles in health and disease, and they may reveal fundamental principles that underlie all host–microbe interactions.
Materials and Methods
Animal Culture and Collection.
Experiments were carried out with H. vulgaris and H. oligactis. H. vulgaris was originally derived from the laboratory of A. Gierer (Max Planck Institute Tübingen, Tübingen, Germany), whereas H. oligactis originally was obtained from P. Tardent (University of Zürich, Zürich, Switzerland). The laboratory animals were cultured at standard conditions at 18°C for >30 years. Free-living animals were isolated from Lake Pohlsee (Schleswig–Holstein, Germany) and Lake Ploen (Schleswig–Holstein, Germany) and cultured for 2 days in filtered water (0.2 μm) from the respective lake, to eliminate digestive food rests before DNA extraction. Species were identified by standard procedure (16).
Molecular Analysis.
For genomic DNA extraction, whole animals were subjected to the DNeasy tissue kit (Qiagen, Hilden, Germany). Universal bacterial PCR primers were used to amplify the region corresponding to positions 27–1492 of the Escherichia coli 16S rRNA gene by using a 30-cycle PCR (17). Resulting PCR fragments were cloned into pGEMT vector (Promega, Madison, WI) and transformed into DH5α E. coli cells (Invitrogen, Karlsruhe, Germany). From each sample, 46 transformants were selected. Plasmid inserts were checked by PCR and subjected to RFLP by using the restriction enzymes HaeIII and Hin6I (Fermentas, Glen Burnie, MD). By sequencing several clones, we confirmed that clones displaying an identical RFLP pattern are members of the same phylotype. Representative plasmids were sequenced by using an LI-COR 4300 DNA analyzer plate sequencer (LI-COR Biosciences, Lincoln, NE). All sequences have been submitted to GenBank (accession nos. EF667892–EF667927). Experimental details are provided in supporting information (SI) Materials and Methods.
Data Analysis.
A total of 68 nonchimeric 16S rDNA sequences were aligned with the ARB software package (18). Phylogenetic analysis revealed 36 phylotypes defined by 97% pairwise sequence identity. A neighbor-joining tree containing these 36 sequences was calculated by using the Olsen correction and a bootstrap resampling of 100 replicates. Details are available in SI Materials and Methods.
Data Analysis with UniFrac.
To test differences between the bacterial communities from each sample, we used the UniFrac computational tool (10). We used the neighbor-joining tree to calculate the fraction of tree branch length unique to any one treatment in pairwise comparisons (the UniFrac metric). The P value for the tree, reflecting the probability that there are more unique branch lengths than expected by chance, was calculated by generating 100 random trees. Choosing the lineage-specific analysis option in the UniFrac computational tool, we tested whether any environment was enriched for particular lineages. The lineage-specific analysis applies the G test of significance to each lineage to determine whether the sequences have a distribution among environments different from the tree overall. The analysis accounted for abundance information resulting from the RFLP analysis. Additionally, we performed UPGMA clustering, by using the weighted UniFrac metric and a jackknife analysis with 100 permutations to access confidence in nodes of the UPGMA tree.
Estimation of Diversity.
The estimation of the number of bacterial phylotypes in each sample was assessed by the Chao1 nonparametric richness estimator implemented in the computational tool EstimateS (version 8, http://purl.oclc.org/estimates). For the purpose of inputting data into the program, we treated each RFLP pattern as a separate sample.
Hoechst Staining and Whole-Cell Hybridization.
Macerates were performed according to the standard protocol (19). Hybridizations of fixed mazerated Hydra cells were done as described by Manz et al. (20) with monofluorescently labeled rRNA-targeted oligonucleotide probes: EUB338 (universal eubacterial probe, positive control) and non-EUB338 (EUB338 antisense probe, negative control) (21). Probes were 5′-end-labeled with either fluorescein (green fluorescence) or Cy3 (red fluorescence). The phylotype-specific oligonucleotide probe HoSym1030 (Cy3, 5′-CCTGTGATAGTCCAGCCG-3′; E. coli positions 1030–1048) was designed by using the probe design function of the ARB software. This probe matched exactly the target region of the 16S rRNA molecule of the dominant phylotype identified in H. oligactis (laboratory). Experimental details are provided in SI Materials and Methods.
Transmission Electron Microscopy.
Experimental details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the members of the T.C.G.B. laboratory for discussion; Friederike Anton-Erxleben for help with scanning electron microscopy; and Ruth Schmitz-Streit, Jens Schröder, Philip Rosenstiel, and Martin Zimmer for valuable comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 617-A1 (to T.C.G.B.).
Abbreviation
- RFLP
restriction fragment length polymorphism.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in the paper have been deposited in the GenBank database (accession nos. EF667892–EF667927).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703375104/DC1.
References
- 1.Steinhoff U. Immunol Lett. 2005;99:12–16. doi: 10.1016/j.imlet.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanmugam M, Sethupathi P, Rhee KJ, Yong S, Knight KL. Inflamm Bowel Dis. 2005;11:992–996. doi: 10.1097/01.mib.0000182869.74648.0f. [DOI] [PubMed] [Google Scholar]
- 4.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFall-Ngai M. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 6.Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, Agata K, Bosch TCG. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch TCG, Krylow SM, Bode HR, Steele RE. Proc Natl Acad Sci USA. 1988;85:7927–7931. doi: 10.1073/pnas.85.21.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennecke T, Gellner K, Bosch TCG. Eur J Biochem. 1998;255:703–709. doi: 10.1046/j.1432-1327.1998.2550703.x. [DOI] [PubMed] [Google Scholar]
- 9.Hemmrich G, Anokhin B, Zacharias H, Bosch TCG. Mol Phylogenet Evol. 2007;44:281–290. doi: 10.1016/j.ympev.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 10.Lozupone C, Knight R. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao A. Scand J Statist. 1984;11:265–270. [Google Scholar]
- 12.Fritsche TR, Horn M, Seyedirashti S, Gautom RK, Schleifer KH, Wagner M. Appl Environ Microbiol. 1999;65:206–212. doi: 10.1128/aem.65.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beninati T, Lo N, Sacchi L, Genchi C, Noda H, Bandi C. Appl Environ Microbiol. 2004;70:2596–2602. doi: 10.1128/AEM.70.5.2596-2602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 15.Rahat M, Dimentman C. Science. 1982;216:67–68. doi: 10.1126/science.7063873. [DOI] [PubMed] [Google Scholar]
- 16.Holstein T. In: Cnidaria: Hydrozoa/Süsswasserfauna von Mitteleuropa. Schwoerbel J, editor. Stuttgart, Germany: Gustav Fisher; 1995. pp. 1–110. [Google Scholar]
- 17.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David CN. Wilhelm Roux's Archiv Entwicklungsmech Org. 1973;171:259–268. doi: 10.1007/BF00577724. [DOI] [PubMed] [Google Scholar]
- 20.Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.