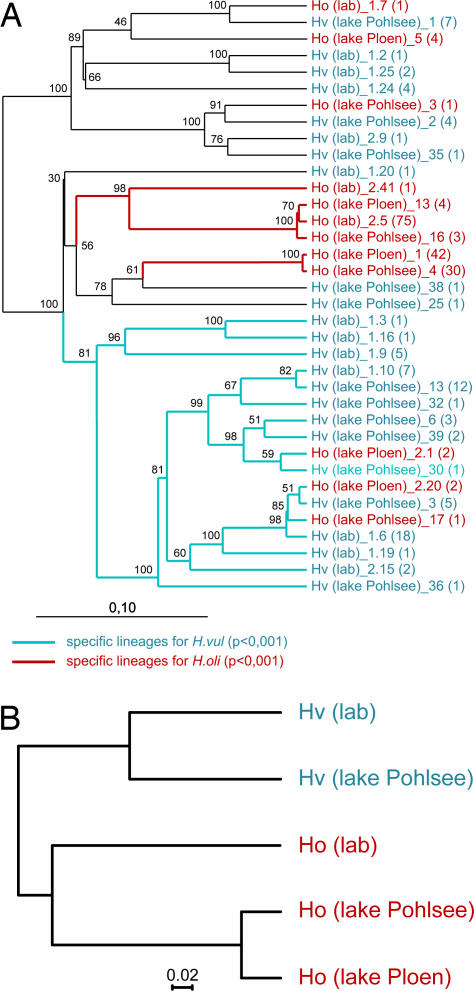
Fig. 3.
Phylogenetic comparison of identified bacterial phylotypes from the different Hydra species. (A) Neighbor-joining tree (Olsen correction) with the 36 identified 16S rDNA phylotypes from five different samples. The number of RFLP patterns within each phylotype is shown in parentheses. Bootstrap values are shown at the corresponding nodes (n = 100). [Scale bar: evolutionary distance (0.1 substitution per nucleotide).] Specific bacterial lineages for the different Hydra species analyzed with UniFrac are indicated in blue (specific for H. vulgaris, P < 0.001) and red (specific for H. oligactis, P < 0.001). (B) Jackknife environment cluster tree (weighted UniFrac metric, based on the 36-sequence tree; ref. 10) of the analyzed bacterial communities. One hundred jackknife replicates were calculated, and each node was recovered with >99.9%. (Scale bar: distance between the environments in UniFrac units.) Hv, H. vulgaris; Ho, H. oligactis; lab, animals from laboratory culture; lake Pohlsee, animals taken from Lake Pohlsee; lake Ploen, animals taken from Lake Ploen.