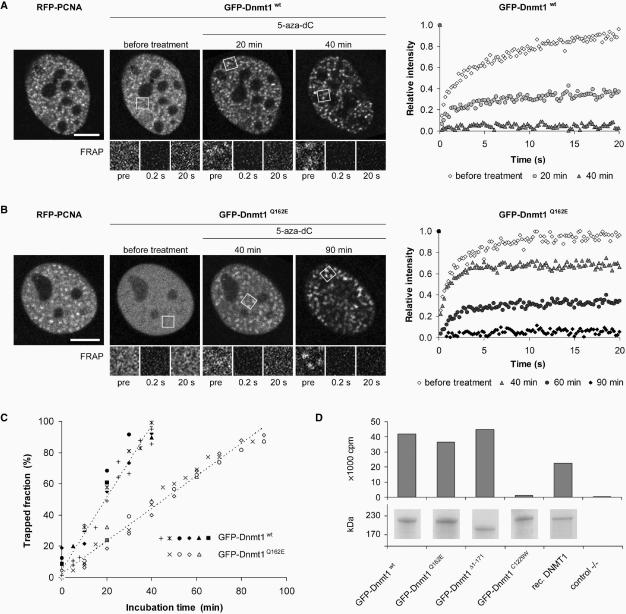
Figure 5.
PCNA-binding-deficient mutant Dnmt1 is catalytically active and shows moderately reduced postreplicative methylation activity in vivo. (A–C) Trapping assay for the analysis of postreplicative methylation activity in vivo. (A, B) Confocal mid sections (upper panel) and corresponding FRAP series (lower panel) of a C2C12 mouse myoblast cell co-transfected with RFP-PCNA and either GFP-Dnmt1wt (A) or GFP-Dnmt1Q162E (B) before and at selected time points after incubation with 5-aza-dC. Before drug treatment the observed cells were in early S-phase as indicated by the RFP-PCNA pattern (left). Boxes indicate bleached ROI's, which are shown magnified in the lower panels at indicated time points of the FRAP series. Quantitative FRAP analysis is shown on the right. Scale bars: 5 µm. (C) Time-dependent increase of immobile fractions of GFP-Dnmt1wt and GFP-Dnmt1Q162E. Individual cells assayed at consecutive time points are indicated by closed (GFP-Dnmt1wt) and open (GFP-Dnmt1Q162E) symbols. Character symbols represent different cells each assayed at a single time point. + symbol indicates data points obtained with C2C12 cells stably expressing GFP-Dnmt1wt (C2C12-GMT1) without co-expression of RFP-PCNA. (D) In vitro methyltransferase assay for GFP-Dnmt1wt, GFP-Dnmt1Δ1-171, GFP-Dnmt1Q162E and the catalytically inactive mutant GFP-Dnmt1C1229W. GFP fusions were expressed in HEK 293T cells, immunopurified and the amount of [3H]CH3 transferred to a hemimethylated oligonucleotide substrate was measured. To normalize for the amount of precipitated protein aliquots were analyzed by SDS-PAGE and Coomassie staining (lower panel).