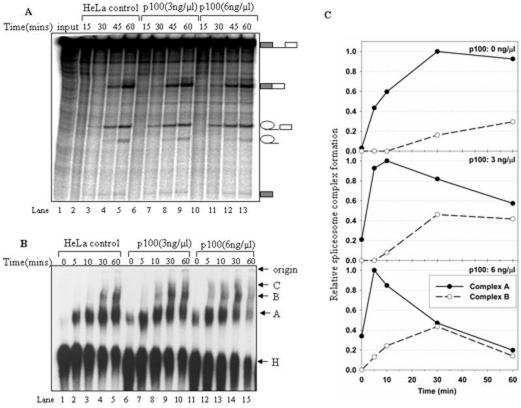
Figure 4.
Effect of p100 protein on the spliceosome assembly and splicing activity. In vitro splicing or spliceosome complex assembly reactions were carried out with AdML splicing substrate in condition indicated above each lane. (A) The kinetics of the splicing reaction following the addition of purified p100 protein. HeLa cell nuclear extracts were first incubated for 20 min on ice in the presence protein p100 storage buffer (lanes 2–5), or different concentrations (3 ng/μl, 6 ng/μl) of p100 protein (lanes 6–13), followed by an addition of ATP and creatine phosphate and a further incubation at 30°C for 10 min. Subsequently, α-32P-labeled AdML pre-mRNA was added, and the incubation continued at 30°C for the time indicated above each lane. RNA products were separated by electrophoresis in denaturing PAGE gels, and visualized by autoradiography. Splicing intermediates and products are indicated on the left. (B) The effect of p100 protein on spliceosomal complex formation in vitro. Native gel analysis of spliceosome complexes formation in a mock-treated splicing reaction (lanes 1–5), or reactions pre-incubated with increasing concentrations (3 ng/μl, 6 ng/μl) of purified p100 protein (lanes 6–15). The gel was visualized by autoradiography. The bands corresponding to the H, A, B and C complexes as well as the gel origin are indicated on the right. (C) Quantitative analysis of spliceosome formation. Native gels were quantified by phosphoimager and normalized by setting the highest A complex value observed within each time series to 1.0.