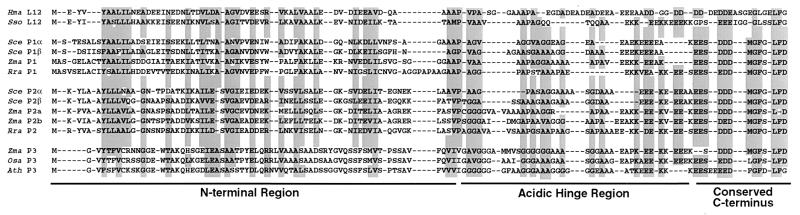
Figure 1.
Alignment of a subset of archaebacterial and eukaryotic 12 kDa P-proteins. The amino acid sequences represent archaebacteria [Sulfolobus solfataricus (Sso), Halobacterium marismortui (Hma)], yeast [Saccharomyces cerevisiae (Sce)], rat (Rattus rattus (Rra)], and plants [Zea mays (Zma), Oryza sativa (Osa), Arabidopsis thaliana (Ath)]. Sequences were aligned using clustal w (49) and were adjusted manually upon visual inspection. Gaps were introduced to ensure maximum homology. Amino acids of conserved physicochemical similarity are shaded based on the following criteria: (i) conserved amino acids must occur in three of the four P-protein groups (i.e., L12, P1, P2, and P3) and (ii) conserved amino acids must occur in at least 75% of the sequences. The N termini, acidic hinge regions, and highly conserved C termini of the 12-kDa P-proteins are indicated.