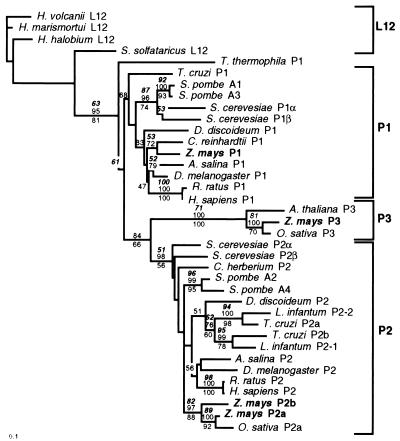
Figure 2.
Amino acid phylogenetic analysis of the 12 kDa P-proteins. A phylogenetic tree was generated by the neighbor-joining method using Kimura-corrected distances in phylip version 3.5c based on the amino acid sequence of 33 eukaryotic 12 kDa P-proteins and 4 archaebacterial L12 proteins. Branch lengths are proportional to the amino acid distances along each branch. Bootstrap values from 500 replicates are indicated. Parsimony bootstrap values for clades supported above the 50% level are indicated below branches, whereas neighbor-joining bootstrap values based on Kimura-corrected distances or uncorrected distances are indicated above the branch (corrected distances are in boldface italics).