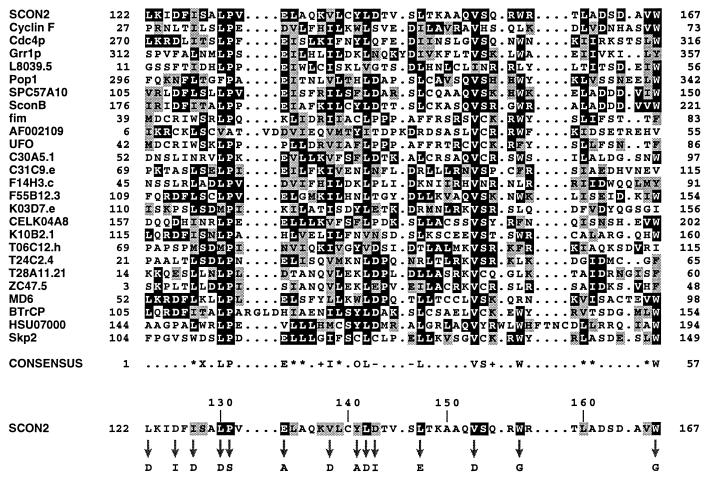
Figure 1.
Sequence analysis of the F-box motif. A representative sample of F-box sequences were globally aligned as described. The F-box proteins included are as follows (GenBank accession numbers are indicated in parentheses): N. crassa SCON2 (U17251); Homo sapiens Cyclin F (Z36714); S. cerevisiae proteins Cdc4p (X05625), Grr1p (M592407), L8039.5 gene product (U19103); Schizosaccharomyces pombe proteins Pop1(Y08391) and unknown protein (Z94864); Emericella nidulans SconB (U21220); Antirrhinum majus fim (S71192); Arabidopsis thaliana hypothetical protein (AF002109) and unusual floral organs (X89224); Caenorhabditis elegans proteins C30A5.1 (L10990), C31C9.e (Z83219), F14H3.c (Z83105), F55B12.3 (Z79757), K03D7.e (Z81562), hypothetical protein (U64849), K10B2.1 (U28730), T06C12.h (Z81116), T24C2.4 (Z68120), T28A11.21 (U80027), ZC47.5 (Z81141); Mus musculus MD6 (X54352); Xenopus laevis β-TrCP (M98268); H. sapiens unknown protein (U07000) and Skp2 (U33761). The numbers bracketing each line of sequence represent the location of each F-box within its parent protein. The most prevalent residue at each position within the alignment is shown as white on black, and similar residues are represented as black on gray. Alignment positions exhibiting less than 20% overall similarity have been left unshaded. The consensus sequence highlights any position within the alignment exhibiting 14 or more occurrences of a single amino acid or class of amino acid (∗, aliphatic; X, hydroxl- or sulfur-containing amino acids and derivatives; +, basic; –, acidic; O, aromatic). The SCON2 F-box sequence has been isolated at the bottom of this alignment to facilitate comparison with the indicated consensus sequence. For this purpose, residues within SCON2 that are conserved with respect to the consensus sequence are represented as black on gray. SCON2 residues that are identical to indicated amino acids in the consensus are shown as white on black. Arrows designate residues within the SCON2 F-box analyzed by site-directed mutagenesis, with corresponding amino acid substitutions indicated below.