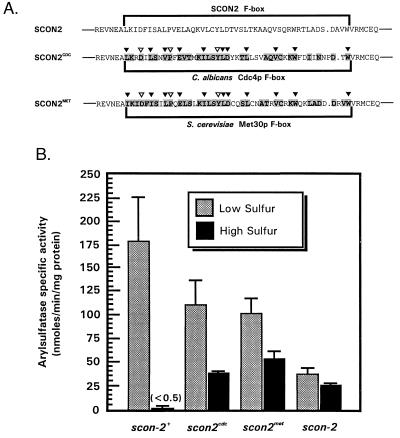
Figure 3.
In vivo functional analysis of chimeric SCON2 F-box mutants. (A) Sequence comparison of SCON2CDC and SCON2MET. SCON2CDC contains a precise replacement of the native SCON2 F-box sequence (encoded within residues 122–167) with the corresponding F-box region from the C. albicans cell cycle regulator Cdc4p (2); SCON2MET features a replacement of this same SCON2 region with the F-box sequence from the S. cerevisiae transcriptional inhibitor Met30p (26). Both chimeric proteins are presented below wild-type SCON2, with F-box domains bracketed within each sequence. F-box sequences were optimally aligned by using clustalw (multiple alignment parameters as previously reported). Residues within SCON2CDC and SCON2MET that are similar or identical to residues within the SCON2 F-box have been printed in bold on a gray background. The C. albicans Cdc4p F-box exhibits 63% similarity and 39% identity with SCON2; the Met30p F-box exhibits 72% similarity and 57% identity to the native SCON2 F-box. Inverted triangles highlight positions within SCON2CDC and SCON2MET corresponding to sites within scon-2+ that already have been studied through site-directed mutagenesis. Specifically, black triangles indicate sites that resulted in sulfur auxotrophy on mutagenesis in scon-2+. (B) Arylsulfatase specific activity in SCON2CDC and SCON2MET. scon2cdc and scon2met strains were assayed for recovery of wild-type sulfur regulation by measuring arylsulfatase enzyme specific activity under low and high sulfur conditions. Full-length scon-2+ sequence as well as the scon-2 mutant strain (10) served as controls. In all cases, reported values indicate the mean arylsulfatase specific activity (in nmols/min per mg of total protein) ± SD from a minimum of five trials of independent homokaryotic transformants. Under high sulfur conditions, scon-2+ transformants displayed an average arylsulfatase activity of 0.37 ± 0.01 nmols p-nitrophenyl/min per mg.