Figure 2.
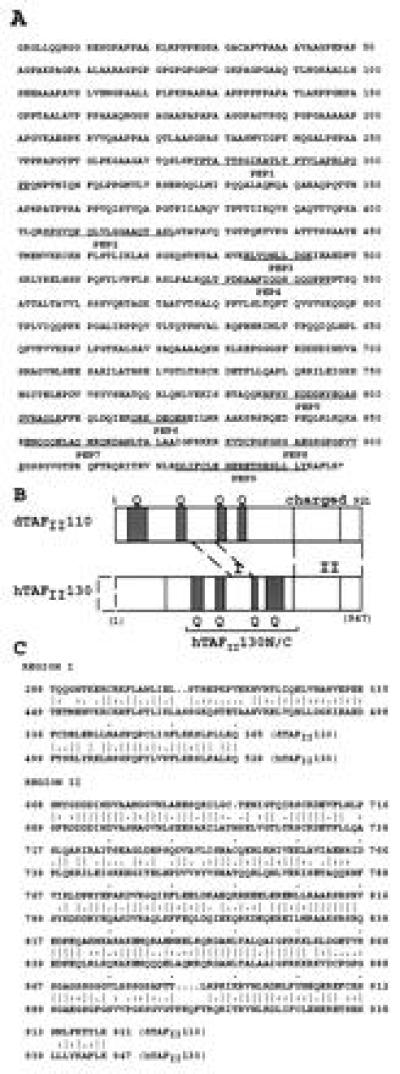
(A) Predicted C-terminal 947 amino acid sequence of hTAFII130. Peptides obtained from microsequencing of immunopurified hTAFII130 protein are underlined. (B) Schematic comparison of hTAFII130 with Drosophila dTAFII110. Region I (residues 449–528): 68% sequence similarity (46% identity) over 80 residues. Region II (residues 689–947): 72% similarity (55% identity) over 259 residues as determined by the bestfit sequence analysis program (Genetics Computer Group, Madison, WI). Glutamine-rich regions (presence of 20–30% glutamines in hTAFII130) are shaded and denoted by Q. hTAFII130N/C subdomain used in protein binding assays is indicated by a bracket. Dotted box represents the predicted missing N-terminal region. (C) Amino acid sequence alignment of conserved residues within regions I and II of dTAFII110 and hTAFII130.
